A new study introduces Molecular Orbital Decomposition and Aggregation (MODA), a quantum-inspired representation (QIR) class descriptor with enhanced predictive capabilities for machine learning models in computational chemistry and materials discovery. MODA incorporates wavefunction information to capture electronic structure intricacies, providing deeper chemical insight and improving performance in unsupervised and supervised learning tasks. It is the first QIR class descriptor capable of distinguishing between intra and intermolecular properties, showing the best performance for intermolecular magnetic exchange coupling predictions among the descriptors tested.
Introduction to Quantum-Inspired Representations (QIR) and Machine Learning (ML)
Machine Learning (ML) is having a significant impact in Quantum Chemistry (QC). Various research fields are benefiting from new computational strategies that combine QC and ML, such as molecular electronics, excited states, low-cost discovery of materials, and catalysis. ML models require data to be transformed into a fixed-size representation, usually in the form of a vector where each element represents a specific attribute or feature. In chemistry, the construction of these elements, typically called descriptors, is particularly challenging due to the diversity and complexity of chemical systems and their interactions.
Development of Descriptors in Chemistry
In the last decade, significant efforts have been made to develop reliable descriptors. These can be classified into three different categories. The first one are cheminformatics descriptors based on either string fingerprints or on descriptive properties that are easily obtainable by a priori knowledge such as the number of aromatic rings or the molecular size. The second category comprises descriptors based on three-dimensional structural information, usually supplemented with parameters inherited from classical mechanics. The third category are descriptors that utilize principles of quantum mechanics to represent the molecular systems and hence can be referred to as quantum-informed representations (QIR).
Quantum-Informed Representations (QIR) and Their Advantages
The main advantage of QIR over classical-informed representations (CIR) descriptors is that the former can explicitly encode the electronic state of a system, including its electronic structure, charge, and spin multiplicity. These attributes are necessary to discriminate, for example, radicals from closed-shell molecules or neutral from charged species, particularly when the molecular geometry is not significantly affected and thus CIR does not significantly change.
Challenges in Predicting Intermolecular Properties
A common problem for both CIR and QIR approaches when combined with ML models is the prediction of intermolecular properties. The majority of the descriptors emphasize the magnitude of short-range interactions to better capture the local atomic environment. As a result, these representations can underperform in cases where medium or long-range, possibly intermolecular interactions are crucial. This issue can be addressed by decoupling intra and intermolecular interactions in the representation and simply ignoring the former in the construction of the descriptor.
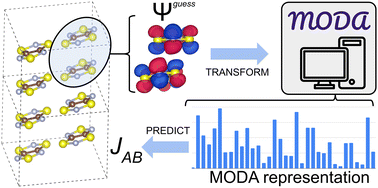
Introduction of Molecular Orbital Decomposition and Aggregation (MODA)
The Molecular Orbital Decomposition and Aggregation (MODA) is introduced as a new QIR descriptor, the first of its kind that allows decoupling strategies. As in Spectrum of Approximated Hamiltonian Matrices (SPA-HM) and Matrix of Orthogonalized Atomic Orbital Coefficients (MAOC), MODA does not require the calculation of self-consistent field (SCF) solutions. Instead, MODA representation can be constructed using well-established guess Hamiltonians, such as the Superposition of Atomic Densities (SAD), the Superposition of Atomic Potentials (SAP), or the extended Huckel method (EH), which are typically starting points in quantum chemistry.
The article titled “Unlocking the Predictive Power of Quantum-Inspired Representations for Intermolecular Properties in Machine Learning” was published on January 1, 2024, in the journal Digital Discovery. The authors of the study are Raul Santiago Piera, Sergi Vela, Mercè Deumal, and Jordi Ribas‐Ariño. The research explores the potential of quantum-inspired representations in predicting intermolecular properties within the field of machine learning. The DOI reference for the article is https://doi.org/10.1039/d3dd00187c.