Medical image segmentation frequently relies on deterministic methods, hindering adaptability to nuanced instructions and limiting the exploration of multiple interpretations. Yuan Lin, Murong Xu, and Marc Hölle, from Friedrich-Alexander-Universität Erlangen-Nürnberg, alongside Chinmay Prabhakar (University of Zurich) and colleagues, address this challenge in their new research, ProGiDiff. They present a novel framework leveraging pre-trained diffusion models , traditionally used for image generation , and adapting them for detailed medical image segmentation, overcoming the need for extensive training datasets. This work is significant because ProGiDiff not only enables natural language prompting for precise control over segmentation, extending beyond simple binary outputs, but also demonstrates impressive cross-modality transferability, successfully adapting to both CT and MR images with minimal retraining.
Medical image segmentation frequently relies on deterministic methods, hindering adaptability to nuanced instructions and limiting the exploration of multiple interpretations
Yuan Lin, Murong Xu, and Marc Hölle, from Friedrich-Alexander-Universität Erlangen-Nürnberg, alongside Chinmay Prabhakar (University of Zurich) and colleagues, address this challenge in their new research, ProGiDiff. They present a novel framework leveraging pre-trained diffusion models, traditionally used for image generation, and adapting them for detailed medical image segmentation, overcoming the need for extensive training datasets. This work is significant because ProGiDiff not only enables natural language prompting for precise control over segmentation, extending beyond simple binary outputs, but also demonstrates impressive cross-modality transferability, successfully adapting to both CT and MR images with minimal retraining. Researchers have identified limitations in existing image generation methods, noting they are primarily deterministic and poorly suited to natural language prompts, struggling to estimate multiple proposals, facilitate human interaction, and enable cross-modality adaptation. Recently, text-to-image diffusion models have demonstrated potential to address this gap; however, training them from scratch necessitates a large dataset, a significant limitation for medical imaging applications.
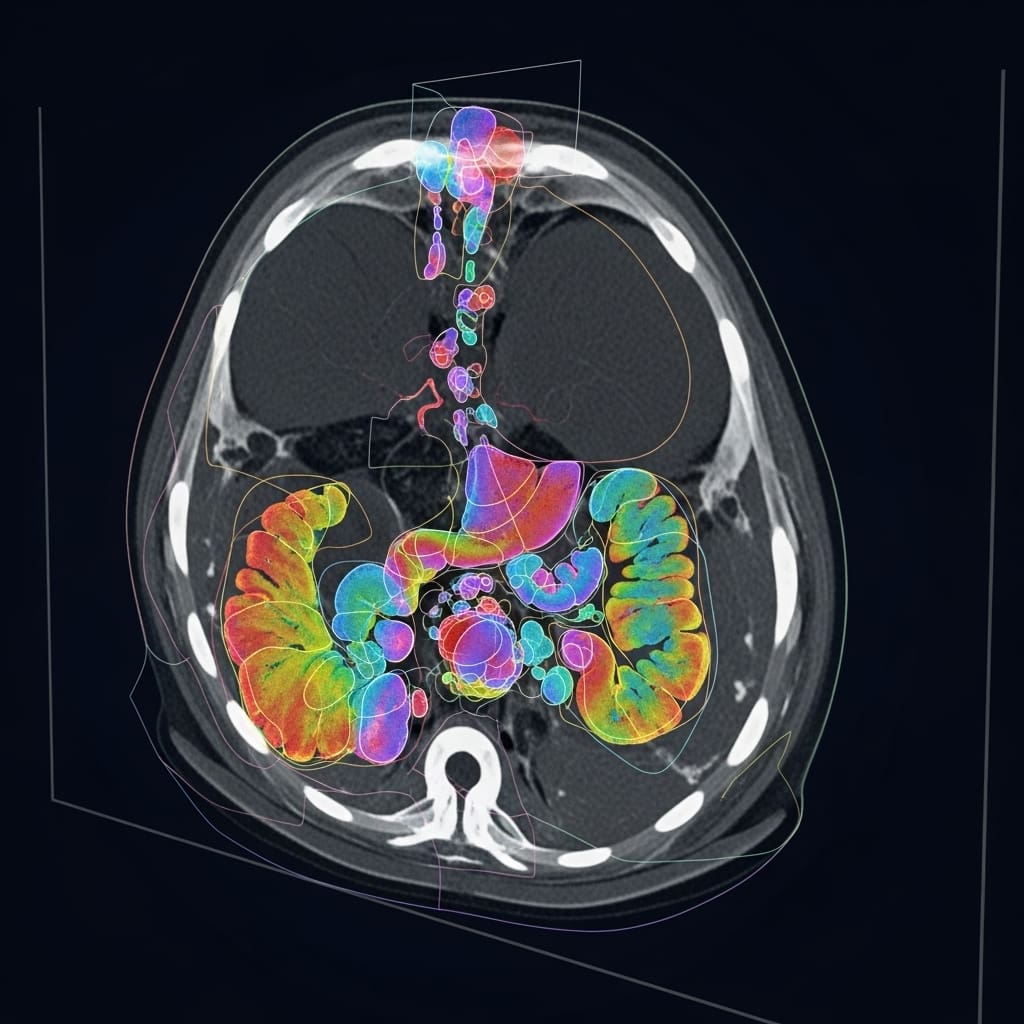
ProGiDiff framework for CT image segmentation offers promising results
Scientists developed ProGiDiff, a novel framework that harnesses pre-trained image generation models for medical image segmentation, addressing key limitations of existing deterministic approaches. The study pioneers a ControlNet-style conditioning mechanism that incorporates a custom encoder for image-based conditioning, enabling precise steering of a pre-trained diffusion model toward the generation of accurate segmentation masks. This design naturally extends to multi-class segmentation by simply prompting the model with the target organ, allowing flexible and scalable anatomical analysis across different clinical scenarios. Experiments conducted on CT images demonstrate that ProGiDiff achieves strong performance compared to prior segmentation techniques, underscoring its potential utility within expert-in-the-loop workflows where multiple segmentation proposals can be iteratively refined and validated.
The conditioning mechanism was carefully engineered to support efficient transfer learning, employing low-rank, few-shot adaptation to enable MR image segmentation and demonstrating robust cross-modality generalization. The methodology centers on embedding semantic prior information into the diffusion process, providing language-driven interpretability and strengthening the alignment between textual prompts and visual medical representations. A custom encoder translates medical image features into representations compatible with the pre-trained diffusion backbone, effectively guiding the segmentation process without requiring retraining of the core model. This strategy enables multi-class segmentation while significantly reducing computational and data requirements.
Furthermore, the research introduces a low-rank adaptation strategy that fine-tunes only a small subset of model parameters, allowing rapid adaptation to new imaging modalities such as MRI. By transferring learned conditioning mechanisms rather than retraining full models, ProGiDiff minimizes dependence on large, modality-specific annotated datasets. Validation experiments on abdominal CT organ segmentation confirm competitive performance, while few-shot domain adaptation experiments on MR images further demonstrate the framework’s generalizability and its promise for broad clinical deployment.
ProGiDiff yields high-accuracy organ segmentation scores
While performance on smaller structures like the pancreas was initially lower, the custom image encoder yielded a 6.09% increase in average Dice score, rising from 70.72% to 75.03%, demonstrating improved feature discriminability in ambiguous regions. This cross-modality adaptation significantly outperforms training from scratch, which required 367.13 million parameters, and exceeds the performance of Diff-UNet at 81.09%. The study also explored a simulated expert-in-the-loop setting, randomly selecting from the top-3 proposals (RandTop-3), and observed consistent gains in average Dice score as the number of generated segmentations increased, reaching approximately 86% with 50 samples. Data shows that ensembling more samples provides limited additional benefit, suggesting a balance between computational cost and performance improvement. These findings underscore the potential of ProGiDiff to enhance clinical workflows and provide clinicians with a range of valid hypotheses for improved diagnostic accuracy.
ProGiDiff excels at CT and MR segmentation, achieving promising results
Scientists have developed ProGiDiff, a novel framework that leverages pre-trained image generation models for medical image segmentation. This approach employs a ControlNet-style conditioning mechanism combined with a custom encoder to guide a diffusion model in generating accurate segmentation masks. By incorporating natural language prompts specifying target organs, ProGiDiff effectively extends segmentation capabilities to multi-class settings without requiring task-specific architectural redesign. Experiments conducted on CT images demonstrate that ProGiDiff achieves strong organ segmentation performance, surpassing prior state-of-the-art methods and highlighting its suitability for expert-in-the-loop clinical systems. The framework further demonstrates adaptability through successful transfer to MR image segmentation using low-rank adaptation (LoRA), requiring only minimal additional training data.
This generative and language-driven approach is particularly well aligned with clinical workflows where expert oversight can iteratively refine and validate multiple segmentation proposals, effectively simulating inter-rater variability and improving robustness. However, the authors note that ensembling an excessive number of generated proposals yields diminishing returns, as increased uncertainty can offset accuracy gains. Future work may focus on identifying optimal ensemble sizes that balance segmentation quality with computational efficiency. Overall, ProGiDiff represents a significant advancement in medical image segmentation by moving beyond purely deterministic pipelines and embracing diffusion-based generative modeling. The ability to condition segmentation outputs through natural language prompts enhances controllability and interpretability, with the potential to streamline clinical workflows and improve diagnostic accuracy. Although the current implementation relies on a pre-trained diffusion model, the demonstrated transferability to MR imaging suggests a promising pathway toward broader applicability in medical domains where labeled data are scarce.