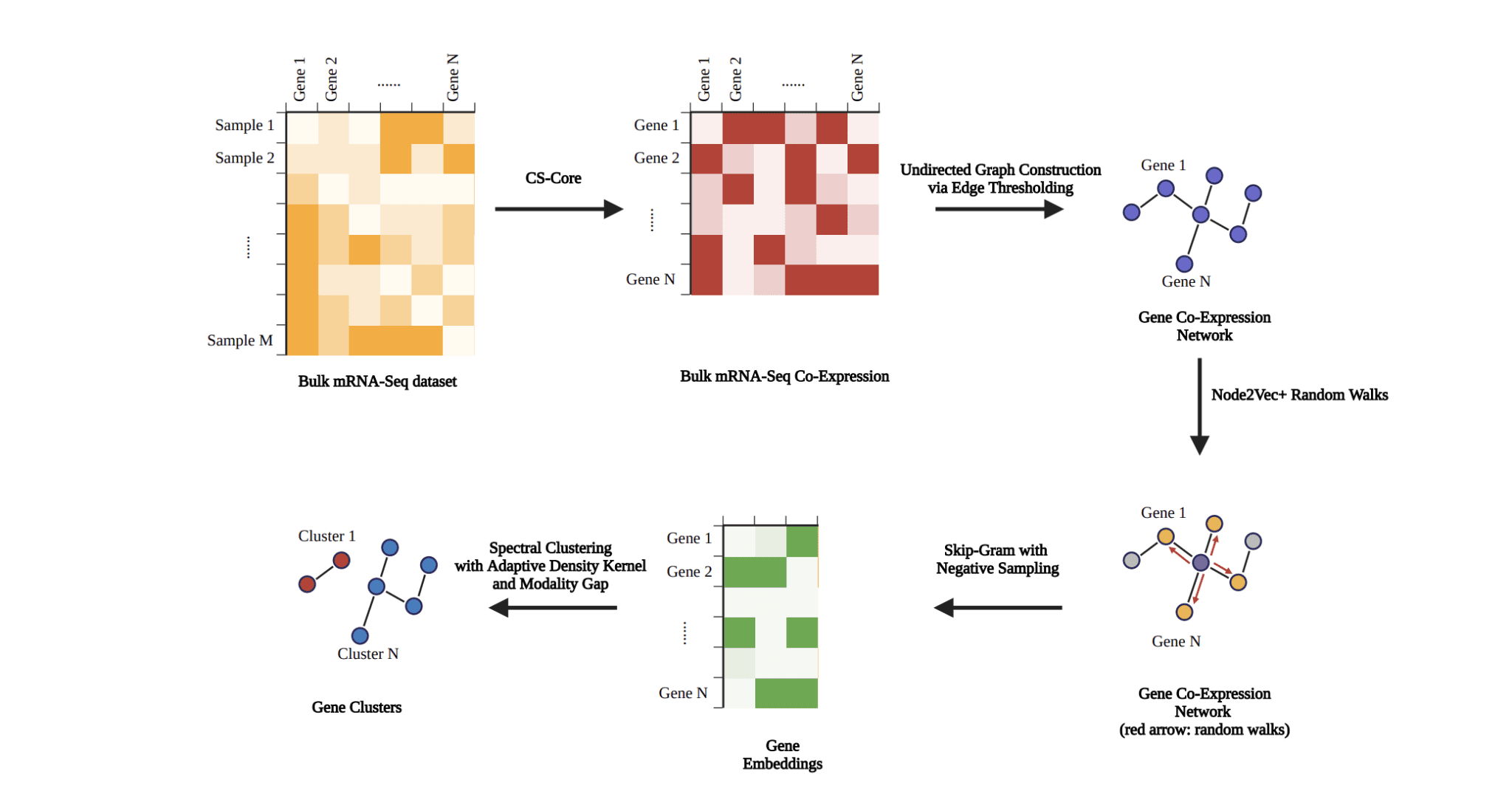
Understanding the genetic basis of disease requires identifying not only individual disease-associated genes, but also the complex relationships between them, and Jake R. Patock from Rice University, Rinki Ratnapriya from Baylor College of Medicine, and Arko Barman from Rice University have developed a new method to achieve this. The team presents a graphical approach that effectively identifies clusters of genes from RNA sequencing data, revealing how genes work together in disease processes. Their technique constructs a network representing gene co-expression, then uses a sophisticated algorithm to map genes into a mathematical space where related genes cluster together, and finally applies a clustering method to identify these groups. This innovative method demonstrates consistent and robust results when applied to data from Age-related Macular Degeneration, and importantly, its design ensures it can be readily applied to a wide range of diseases, accelerating the discovery of underlying genetic mechanisms and potential therapeutic targets.
Genomics, Networks, and Machine Learning Integration
This compilation of research explores the intersection of genomics, machine learning, and network analysis, with a particular focus on age-related macular degeneration and cancer. The collection highlights a growing trend towards integrating diverse data types, such as transcriptomics and genomics, to gain a more comprehensive understanding of biological processes. Machine learning techniques, including clustering and deep learning, are heavily employed to analyze genomic data, identify patterns, and predict outcomes. Network analysis emerges as a powerful approach for modeling biological systems, identifying key genes and proteins, and understanding their interactions through techniques like co-expression network inference.
Several studies specifically utilize single-cell RNA sequencing, demonstrating increasing interest in understanding cellular heterogeneity. Researchers frequently employ dimensionality reduction and clustering techniques to simplify and organize complex genomic data. A growing emphasis on explainable AI indicates a desire to create machine learning models that are more interpretable and transparent.
Gene Embeddings Reveal Disease-Associated Clusters
Scientists developed a novel graph-based method for identifying gene clusters associated with disease, employing RNA-Seq data and robust clustering analysis. The study begins by constructing a gene co-expression network, representing relationships between genes based on their expression patterns. Researchers then leveraged the Node2Vec+ algorithm to compute gene embeddings, creating numerical representations of each gene within the network that capture its contextual relationships with other genes. These embeddings, representing genes in a multi-dimensional space, facilitate the identification of functionally similar genes.
Following the creation of gene embeddings, the team implemented spectral clustering to identify distinct groups of genes. This technique partitions genes into clusters based on the distances between their embeddings, revealing potential functional relationships and shared biological pathways. To ensure the stability and optimality of the entire process, researchers jointly optimized all steps using a Tree-structured Parzen Estimator, refining parameters and maximizing the reliability of the resulting clusters. The work begins by constructing a gene co-expression network, mapping relationships between genes, and then computes gene embeddings, representing each gene as a point in a high-dimensional space that captures its expression patterns and connections within the network. Finally, the team clustered these gene embeddings to reveal groups of genes with similar expression profiles, potentially indicating shared functions or pathways involved in the disease. The core breakthrough lies in a joint optimization process, where network construction, embedding computation, and clustering are simultaneously refined to maximize overall performance.
This contrasts with traditional approaches that optimize each step individually. The team’s cost function focuses on the quality of the final clustering, ensuring superior performance. The team constructed a gene co-expression network from RNA sequencing data and then used a sophisticated algorithm, Node2Vec+, to compute gene embeddings, effectively representing genes within a network context. Subsequent spectral clustering of these embeddings successfully identified groups of genes, revealing potential functional relationships. The method demonstrates the ability to consistently generate robust gene clusters from RNA-Seq data, offering a valuable tool for understanding complex disease mechanisms. By grouping known AMD-related genes, the approach can help pinpoint shared biological pathways and potentially uncover previously unrecognized genes involved in the disease. This clustering strategy moves beyond focusing on individual genes, aiming to identify underlying disease mechanisms for more effective therapeutic interventions.
👉 More information
🗞 A Graphical Method for Identifying Gene Clusters from RNA Sequencing Data
🧠 ArXiv: https://arxiv.org/abs/2511.09590