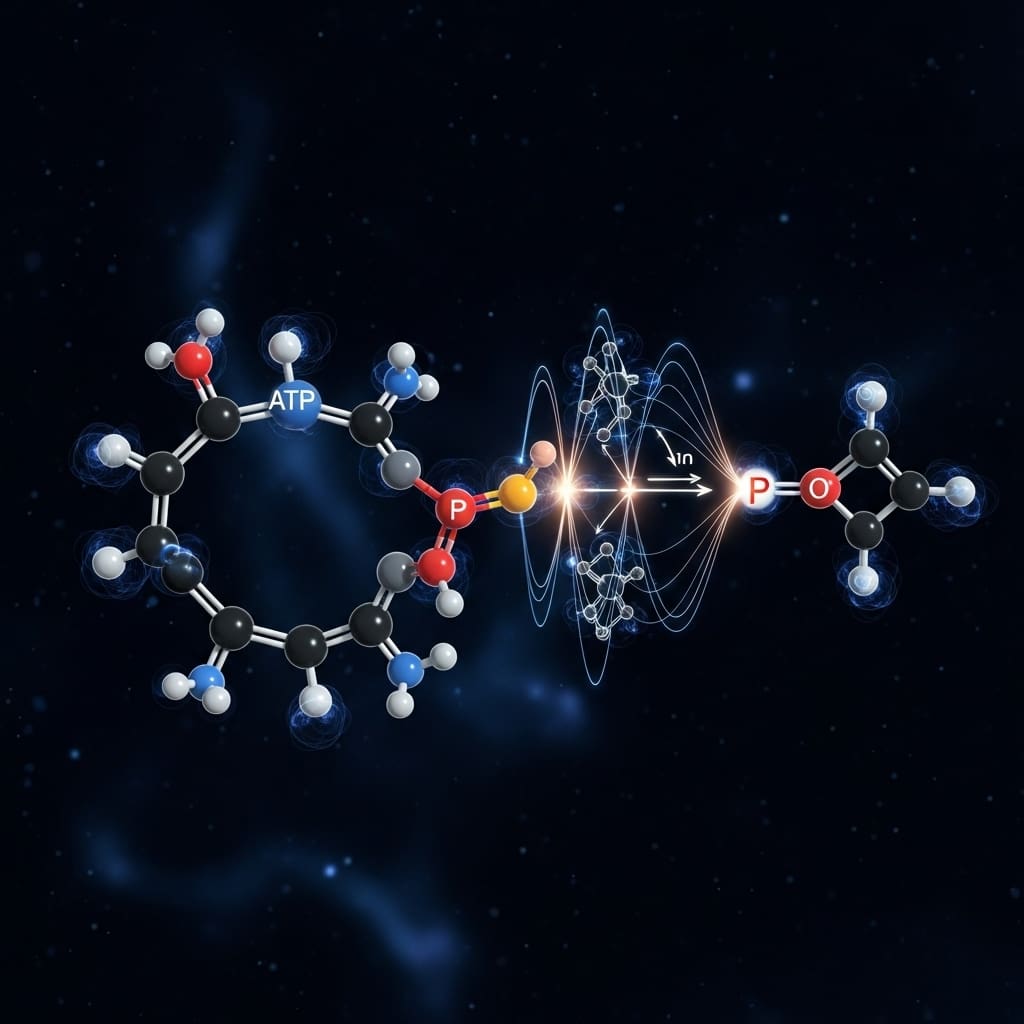
Researchers have begun to quantify the computational cost of simulating fundamental biochemical processes using emerging quantum algorithms. Ryan LaRose (Michigan State University), Alan Bidart (Brown University), and Ben DalFavero (Michigan State University), alongside Economou et al., investigated the resource demands of modelling ATP/metaphosphate hydrolysis , a crucial reaction underpinning metabolism, cellular signalling, and potential cancer treatments. Their work, utilising exact classical simulation, numerical estimation and analytical bounds, represents a significant step towards determining the feasibility of applying quantum computers to real-world biological problems. By comparing the eigensolver, Krylov, and phase estimation methods, the team demonstrate that heuristic approaches may offer a pathway to tackling these complex simulations on current and near-future quantum hardware, and have generously provided their complete dataset and code as benchmarks for further development.
By comparing the eigensolver, Krylov, and Phase estimation methods, the team demonstrate that heuristic approaches may offer a pathway to tackling these complex simulations on current and near-future quantum hardware, and have generously provided their complete dataset and code as benchmarks for further development.
Quantum Algorithms For ATP Hydrolysis Analysis
This work extends beyond simply analysing algorithms in isolation; it delves into the practical overhead costs associated with specific problems and hardware limitations, a crucial step towards realising tangible quantum advantages. The study unveils a detailed comparison of these three algorithms, categorised by their relevance to different eras of quantum computing: noisy intermediate-scale quantum (NISQ), near-future “MegaQuop” devices, and fault-tolerant application-scale quantum (FASQ) computing. Researchers meticulously analysed resources both generally and specifically compiled for particular quantum hardware platforms, utilising both theoretical results and state-of-the-art quantum compilation methods. This finding is significant as it identifies a pathway for achieving meaningful results with existing and emerging quantum technologies.
Experiments show that the team’s approach extends beyond asymptotic scaling laws, providing a more accurate picture of end-to-end quantum protocol costs when factoring in problem specifics and hardware constraints. They focused on magnesium-coupled metaphosphate hydrolysis, a problem previously unaddressed in quantum resource estimation literature, highlighting its importance in biochemistry, biology, and human health. The research establishes that this problem is amenable to treatment across all three quantum computing eras, NISQ, MegaQuop, and FASQ, through classical pre-processing and innovative methods. This open-source approach fosters collaboration and accelerates progress in the field. The work opens exciting possibilities for simulating complex biochemical processes with unprecedented accuracy, potentially leading to breakthroughs in drug discovery, metabolic engineering, and our understanding of fundamental life processes. The detailed resource estimations and benchmarks provided in this study will undoubtedly serve as a valuable resource for the quantum computing and biochemistry communities alike.
Quantum Algorithm Benchmarking for Biomolecular Energy Estimation
Researchers employed exact classical simulation alongside numerical estimation and analytical bounds to rigorously assess the feasibility of these algorithms on both current and near-future quantum devices. The team engineered a Hamiltonian downfolding technique to reduce the complexity of the biomolecular Hamiltonians, enabling more efficient quantum simulations. This involved systematically reducing the size of the Hamiltonian while preserving essential physics, thereby lowering the demands on quantum hardware. Furthermore, scientists developed an ADAPT-VQE approach, a tailored variational quantum eigensolver implementation, to optimise the selection of basis states and minimise circuit depth.
Crucially, the study pioneered a quantum subroutine for time evolution, leveraging advanced techniques to accurately simulate the dynamics of the biochemical reaction. Experiments employed classical simulation to validate and benchmark the performance of the quantum algorithms, utilising high-performance computing resources to model the behaviour of the system. Quantum Krylov methods were analysed with and without thresholding, allowing researchers to quantify the impact of noise reduction techniques on algorithm accuracy and resource consumption. To assess the scalability of the algorithms, the team established rigorous bounds for quantum phase estimation, providing theoretical limits on the required quantum resources. A key methodological innovation was the implementation of circuit compilation and compression techniques, designed to optimise quantum circuits for specific hardware platforms. This involved mapping the logical quantum gates onto the physical qubits of the target device, minimising gate count and coherence time requirements.
Variational algorithms minimise quantum resource needs by leveraging
Experiments revealed that the energy of the metaphosphate hydrolysis reaction, as measured by classical electronic structure theories like CCSD and CAM-B3LYP, varies significantly along the reaction coordinate, particularly at the transition state, highlighting the quantum mechanical complexity of the problem. Measurements confirm that the chosen biochemical problem is feasible for treatment across different quantum computing eras, NISQ, MegaQuop, and FASQ, due to classical pre-processing and other methods employed. The team meticulously analysed resources both generally and specifically compiled for particular quantum hardware platforms, utilising theoretical results, numerical simulation, and state-of-the-art quantum compilation techniques. Data shows that the energy of the reaction, as determined by CCSD(T), differs most significantly from other methods at the transition state, indicating the difficulty in accurately modelling this critical point. The breakthrough delivers a valuable resource for the quantum computing community, providing both data and code to facilitate further research and development in this rapidly evolving field.
Quantum Heuristics Outperform For Biomolecular Energy Estimation
The authors acknowledge limitations stemming from the inherent challenges in accurately predicting resource needs for complex quantum protocols, particularly when factoring in specific hardware and software details. Future research could focus on refining these estimations and exploring the potential of these algorithms on increasingly powerful quantum computers.
👉 More information
🗞 The cost of quantum algorithms for biochemistry: A case study in metaphosphate hydrolysis
🧠 ArXiv: https://arxiv.org/abs/2601.19059