Designing new drug molecules requires navigating an immense landscape of possible structures, a process that remains a significant hurdle in pharmaceutical research. Aaron Mark Thomas, Yu-Cheng Chen, and colleagues from the University of Birmingham and QunaSys, along with Hubert Okadome Valencia, Sharu Theresa Jose, and Ronin Wu, present a new approach to this challenge with QCA-MolGAN, a generative model that leverages the principles of quantum computing. This innovative system combines quantum circuits with a generative adversarial network, effectively learning the characteristics of promising drug candidates and aligning them with desired properties. The team further enhances this process by incorporating a multi-agent reinforcement learning network, which simultaneously optimises crucial metrics like drug-likeness, solubility, and ease of synthesis, ultimately generating molecules with a more balanced and desirable profile than previous methods.
GANs and Quantum Chemistry for Molecules
This research explores a new approach to generative chemistry, aiming to design and create novel molecules with specific, desired properties. The team investigates integrating quantum computing with deep learning methods to accelerate drug discovery and materials science, addressing a key challenge of limited diversity in generated molecules. The work focuses on Generative Adversarial Networks (GANs), where a generator creates molecules and a discriminator evaluates their quality. Researchers developed the Quantum Associative Adversarial Network (QAAN), employing quantum associative memory to improve the generator’s ability to explore chemical space and avoid suboptimal solutions.
Quantum Boltzmann Machines are also investigated as components within this framework, and a technique called Simultaneous Perturbation Stochastic Approximation optimises the quantum components. The models optimise for multiple molecular properties simultaneously, such as solubility, synthetic accessibility, and drug-likeness, representing molecules using graph neural networks to capture their structure and properties. An improved GAN training technique, the Wasserstein GAN, stabilises the training process, and metrics like the Synthetic Accessibility Score and Quantitative Estimate of Drug-likeness evaluate the generated molecules. Results demonstrate that these hybrid quantum-classical models, particularly QAAN, generate more diverse sets of molecules compared to traditional GANs, enhancing the exploration of chemical space. While scaling up the quantum components remains a challenge, the research represents a significant step towards more efficient molecular design.
Quantum and Classical GAN for Molecular Design
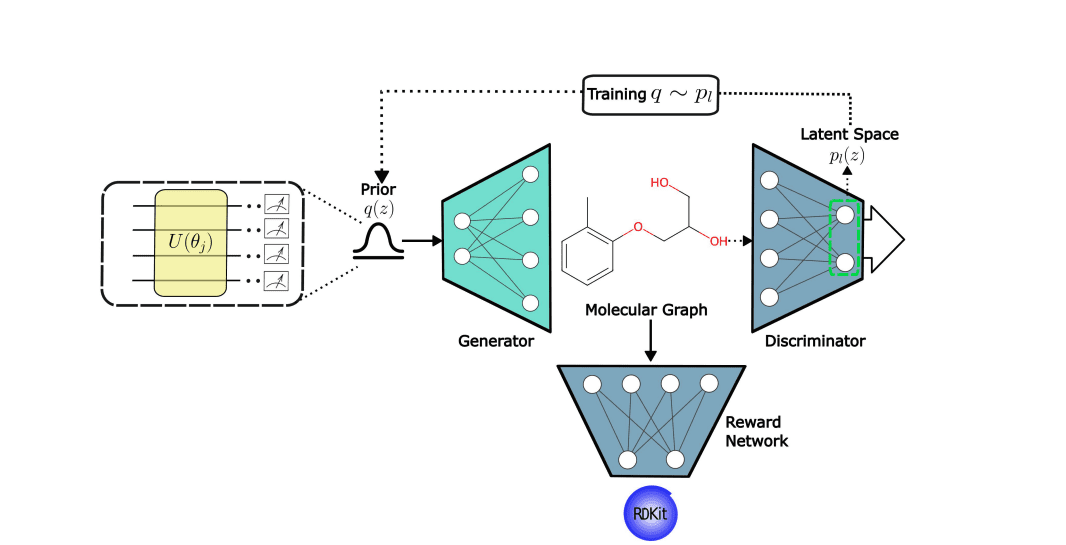
Scientists engineered a novel framework, Quantum Circuit Associative MolGAN (QCA-MolGAN), to accelerate the design of new molecules with targeted properties. This innovative approach uniquely combines the strengths of quantum computing and classical machine learning, addressing limitations in existing drug discovery methods. The framework uses a quantum circuit Born machine (QCBM) as a probabilistic model, generating latent samples for molecular design, while a classical generative adversarial network (GAN) refines these structures into viable drug candidates. The team trained the QCBM to align with the internal representations of the GAN’s discriminator, creating an associative memory within the system.
This associative training enhances the diversity of generated molecules, overcoming the problem of mode collapse seen in traditional GANs. To further refine molecular properties, the researchers incorporated a multi-agent reinforcement learning (MARL) network, employing specialized agents to predict and optimise specific molecular properties, such as quantitative estimate of drug-likeness, octanol-water partition coefficient, and synthetic accessibility. Experiments on the QM9 dataset demonstrate the superior performance of this approach, achieving improved macro averages across all key properties, showcasing the potential for designing molecules with tailored functionalities.
Quantum GANs Design Novel Drug Molecules
Scientists have developed a new framework, Quantum Circuit Associative MolGAN (QCA-MolGAN), to accelerate the design of novel drug molecules with targeted properties. This innovative approach combines the power of quantum computing with classical machine learning techniques, addressing limitations found in existing generative models. The team integrated a quantum circuit Born machine (QCBM) to serve as a probabilistic model, providing latent samples to a classical generative adversarial network (GAN). Crucially, the QCBM is trained to align with the internal workings of the GAN’s discriminator, creating a powerful associative memory that enhances the diversity of generated molecules.
The research team further enhanced QCA-MolGAN by incorporating multi-agent reinforcement learning (MARL), employing specialized agents to independently predict and optimise distinct molecular properties. These agents work collaboratively to guide the molecular generation process, simultaneously optimising key metrics such as quantitative estimate of drug-likeness, octanol-water partition coefficient, and synthetic accessibility scores. Experiments on the QM9 dataset demonstrate the superiority of QCA-MolGAN, achieving improved performance across all key properties when compared to existing methods. This hybrid quantum-classical approach overcomes challenges associated with scaling quantum models and preventing mode collapse in GAN architectures, offering a promising pathway for the efficient discovery of new drug candidates and materials with tailored properties.
Quantum Generation of Optimised Drug Candidates
The research team presents QCA-MolGAN, a new computational framework that combines quantum and classical techniques to generate potential drug molecules with desired characteristics. This approach utilises a quantum circuit Born machine as a guiding prior, effectively shaping the generation process to prioritise molecules exhibiting specific properties like drug-likeness, solubility, and synthetic accessibility. Experiments on a standard chemical dataset demonstrate that QCA-MolGAN successfully enhances these targeted properties in the generated molecules, while also maintaining their validity and novelty. Notably, the integration of a multi-agent reinforcement learning strategy further improved the balance of these chemical properties across the generated molecules. The authors acknowledge a limitation in the model’s performance, observing some degree of ‘mode collapse’ where the generator focuses on a limited range of high-reward samples, although these samples remain diverse from one another. Future work will focus on refining the multi-objective optimisation process, potentially through dynamic reward weighting, and improving training stability with a carefully managed hyperparameter schedule.
👉 More information
🗞 QCA-MolGAN: Quantum Circuit Associative Molecular GAN with Multi-Agent Reinforcement Learning
🧠 ArXiv: https://arxiv.org/abs/2509.05051