Researchers are increasingly applying graph neural networks to protein structure prediction, yet current methods struggle with both capturing information across multiple scales and efficiently modelling long-range dependencies. Shih-Hsin Wang, Yuhao Huang, and Taos Transue, from the University of Utah’s Department of Mathematics and Scientific Computing and Imaging (SCI) Institute, alongside Justin Baker (UCLA) and Jonathan Forstater and Thomas Strohmer (UC Davis), address these limitations with a novel multiscale graph-based learning framework. Their approach constructs a hierarchical graph, combining detailed subgraphs representing secondary structural motifs with a coarse-grained graph illustrating spatial arrangement, and utilises dedicated graph neural networks for both local and global feature learning. This modular design not only preserves maximal expressiveness, ensuring no loss of vital information, but also demonstrably improves prediction accuracy and reduces computational demands across established benchmarks, representing a significant advance in the field.
Existing graph-based methods struggle with efficiently capturing both detailed local interactions and long-range dependencies within proteins, hindering their ability to model complex biological systems.
This research introduces a hierarchical graph representation that combines fine-grained subgraphs corresponding to secondary structural motifs, such as alpha-helices and beta-strands, with a coarse-grained graph connecting these motifs based on their spatial arrangement. The framework employs two distinct graph neural networks, one operating within each motif to capture local interactions and another modeling relationships across motifs at a higher level.
This modular design allows for flexible integration of various GNN architectures, enhancing adaptability and performance. Theoretically, the researchers demonstrate that this hierarchical framework preserves maximal expressiveness, ensuring no critical structural information is lost during processing.
Empirical results across multiple benchmarks reveal that integrating baseline GNNs into this multiscale framework significantly improves prediction accuracy while simultaneously reducing computational demands. The innovation addresses a key challenge in protein modeling: representing proteins at a residue level that balances computational efficiency with the need to capture crucial multiscale features.
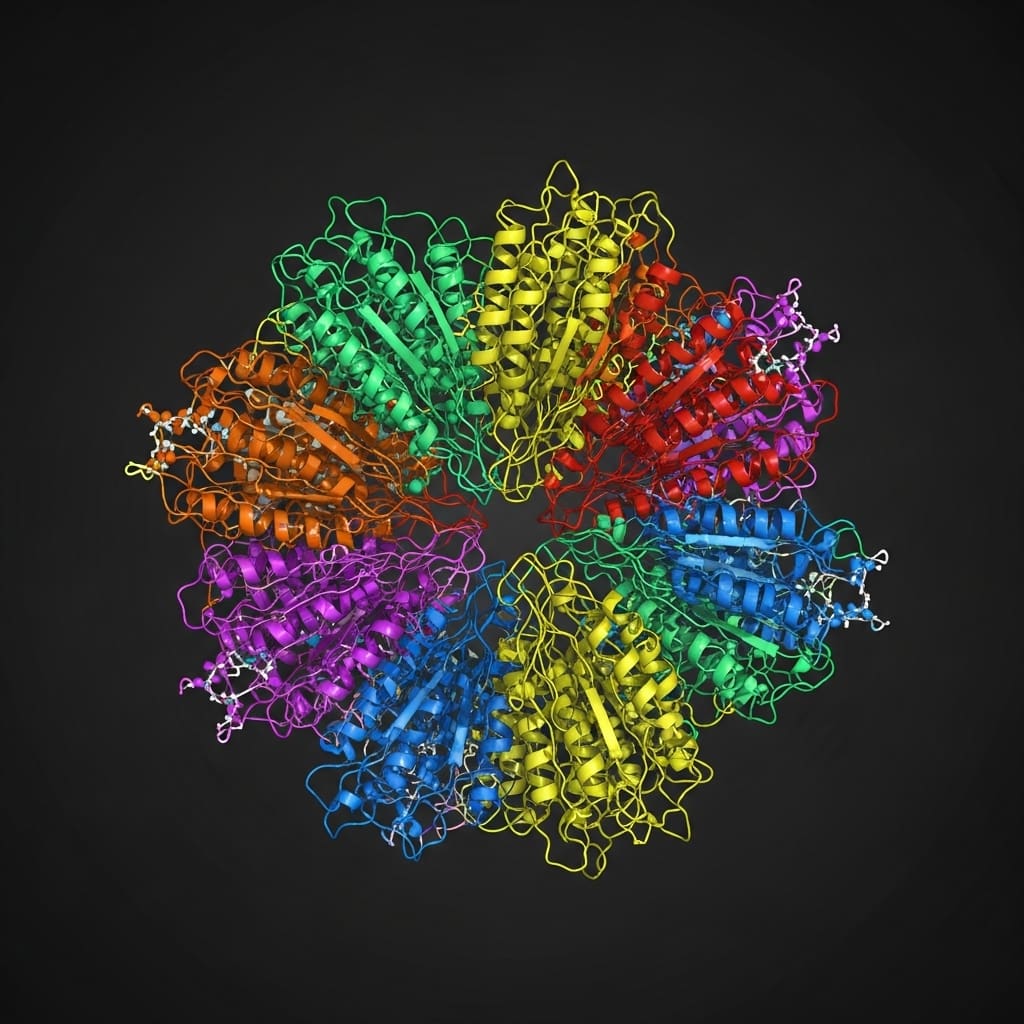
Proteins, fundamental building blocks of life, often exhibit distinct structural changes without alterations to their primary amino acid sequence, impacting their function and potentially leading to disease. A prime example is the prion protein, which can misfold from a normal, helical form to a pathogenic, beta-sheet-rich structure, causing fatal neurodegenerative diseases.
Current residue-level approaches often overlook these critical secondary structures, limiting their ability to differentiate between biologically distinct protein states. This new framework explicitly incorporates these secondary structures into the graph representation, enabling more accurate modeling of protein folding and function.
The proposed framework constructs a sparse, geometry-aware graph, leveraging domain expertise to segment protein sequences into motifs and establish a hierarchical structure. This approach not only enhances scalability and efficiency but also preserves geometric fidelity, crucial for understanding protein interactions. By combining fine-grained local interactions with higher-level structural relationships, the researchers have created a powerful tool for advancing computational protein modeling and potentially accelerating drug discovery and disease research.
Construction of a multi-scale protein structure graph and feature learning with dual graph neural networks enables improved protein function prediction
A hierarchical graph representation forms the core of this research, designed to efficiently model protein structures at multiple scales. The methodology begins by constructing fine-grained subgraphs, each representing a secondary structure motif such as α-helices, β-strands, and loops, with residues within these motifs serving as individual nodes.
Simultaneously, a single coarse-grained graph is created, connecting these motifs based on their spatial arrangement and relative orientation within the overall protein structure. Two distinct graph neural networks are then employed for feature learning within this framework. The first GNN operates independently on each of the fine-grained motif subgraphs, capturing local interactions and generating motif-level embeddings.
These learned embeddings are subsequently used to construct the coarse-grained inter-structural graph, where each motif is represented as a single node. A second GNN then performs message passing on this coarse-grained graph, modelling higher-level relationships between motifs and ultimately generating global features for protein structure prediction.
This modular design allows for flexible integration of different GNN architectures at each stage, enabling exploration of various feature learning strategies. The theoretical work demonstrates that this hierarchical framework preserves maximal expressiveness, preventing any loss of critical structural information during the multiscale representation process. Empirical validation across multiple benchmarks confirms that this approach improves prediction accuracy and reduces computational cost when compared to baseline GNN methods.
Secondary structure motifs enhance protein representation via hierarchical graph networks by capturing local and global dependencies
Researchers developed a hierarchical graph-based learning framework that achieves substantial improvements in protein representation learning with reduced model sizes. The core of this work lies in a two-level graph construction, utilizing secondary structures as high-level motifs to capture both local geometry and long-range dependencies.
This design demonstrably preserves maximal expressiveness, ensuring no critical information is lost during processing. The proposed framework constructs a hierarchical graph representation comprising fine-grained subgraphs corresponding to secondary motifs, such as α-helices, β-strands, and loops, alongside a single coarse-grained graph connecting these motifs based on spatial arrangement and relative orientation.
Two graph neural networks are employed for feature learning, with the first operating within individual secondary motifs to capture local interactions. The second GNN models higher-level relationships across these motifs, enabling efficient processing of complex protein structures. Theoretical analysis confirms that this hierarchical framework maintains the desired maximal expressiveness, a critical property for accurate protein representation.
Specifically, a maximally expressive GNN with a depth of one layer can distinguish between the attributed SCHull graphs of any two non-isomorphic generic point clouds, assuming the point coordinates are algebraically independent over the field of rational numbers. This condition holds true for the majority of protein structures examined in the study.
Local frames, orthogonal matrices defining local 3D coordinate systems, are equivariant under rotations and reflections, further enhancing the robustness of the model. The study leverages these local frames to ensure consistent and accurate representation of protein geometry. This biologically grounded hierarchical design offers provable sparsity and expressiveness guarantees while maintaining high computational efficiency, representing a significant advancement in multiscale graph-based modeling.
Hierarchical graph networks enhance protein structure prediction through multiscale representation learning
Researchers have developed a new multiscale graph-based learning framework that improves the accuracy and efficiency of protein structure modelling. This framework addresses limitations in existing graph neural networks by effectively capturing both local interactions within secondary structural motifs and long-range dependencies between them.
The approach constructs a hierarchical graph representation, combining detailed subgraphs representing elements like alpha-helices and beta-strands with a broader graph illustrating their spatial relationships. This modular framework utilises two graph neural networks operating at different scales, one focusing on interactions within motifs and the other on relationships between them.
Theoretical analysis confirms that this hierarchical structure maintains maximal expressiveness, preventing information loss during processing. Empirical results demonstrate significant improvements in accuracy and reductions in computational cost when integrating existing graph neural networks into this multiscale framework across several benchmark datasets.
The authors acknowledge that future work could explore broader applications and refinements of the model. They highlight the potential for further research into alternative graph neural network architectures and the investigation of different methods for constructing the hierarchical graph representation.
While the current work demonstrates strong performance on established benchmarks, the generalizability to entirely novel protein structures remains an area for continued investigation. The framework’s computational efficiency and scalability with larger datasets also warrant further study to ensure its practical applicability to increasingly complex biological systems.
👉 More information
🗞 Towards Multiscale Graph-based Protein Learning with Geometric Secondary Structural Motifs
🧠 ArXiv: https://arxiv.org/abs/2602.00862