Capturing the rapid, three-dimensional dynamics of a beating heart presents a significant challenge for conventional imaging techniques, which often struggle to balance spatial resolution with the necessary speed. Yi Gong, Xinyuan Zhang, and Jichen Chai, from The University of Texas at Dallas, alongside Yifei Lou from The University of North Carolina at Chapel Hill, and their colleagues, now address this limitation with a new imaging framework that combines the strengths of light-sheet microscopy and compressed sensing. Their approach achieves efficient, low-damage cardiac imaging by acquiring fluorescence signals using a coded light pattern, and then reconstructing detailed images using a flexible ‘plug-and-play’ algorithm. This method successfully preserves fine structural details in rapidly changing tissue, delivering clear images even with significant data compression, and represents a substantial advance for high-speed, low-light biological imaging.
Zebrafish Heart Imaging with Light-Sheet Microscopy
Scientists are pioneering advanced microscopy techniques to visualize dynamic biological processes, with a particular focus on the heart, using zebrafish as a model organism. This research combines innovative hardware, such as light-sheet microscopy, with sophisticated image reconstruction and analysis algorithms, often employing deep learning techniques, to observe the heart’s intricate workings with unprecedented clarity. Zebrafish are ideal for this research because their early development allows for easy visualization of internal organs, and their hearts share structural similarities with mammalian hearts. Light-Sheet Microscopy illuminates the sample with a thin sheet of light, reducing blurring and minimizing harmful light exposure. Deep learning algorithms play a crucial role in removing noise from images, reconstructing three-dimensional volumes, and identifying specific features within the heart, such as chambers and muscle fibers. Techniques like compressed sensing and snapshot imaging further accelerate the process by reducing the amount of data needed for reconstruction.
Compressive Light-Sheet Microscopy for Rapid Cardiac Imaging
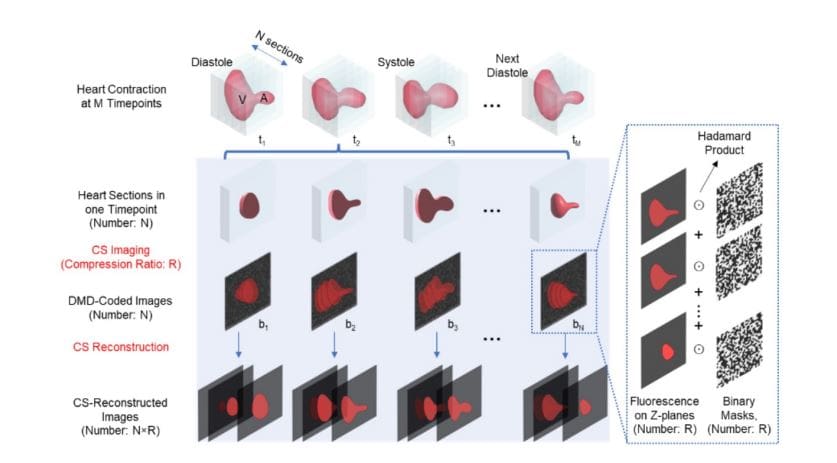
Scientists engineered a high-performance imaging system that integrates Compressive Sensing with Light-Sheet Microscopy to capture rapid cardiac dynamics with exceptional clarity and minimal phototoxicity. The system employs a Digital Micromirror Device to perform compressed acquisition of fluorescence signals, enabling multi-slice imaging with high temporal precision at frame rates reaching 200 volumes per second while maintaining cellular resolution. This hardware design significantly enhances imaging speed without compromising resolution or sample viability, and reduces required storage. To reconstruct images from encoded signals, the team developed a novel reconstruction framework incorporating a Plug-and-Play algorithm solved using the alternating direction method of multipliers, which flexibly integrates advanced denoisers including Tikhonov, Total Variation, and BM3D. The method further introduces temporal regularization, enforcing smoothness between adjacent z-slices to preserve structural continuity during dynamic imaging, effectively mitigating artifacts and resulting in high-quality reconstructions. Experiments performed on zebrafish hearts demonstrate the effectiveness of this methodology, successfully reconstructing cellular structures even under high compression ratios, providing a powerful tool for investigating cardiac function with unprecedented spatiotemporal fidelity.
Rapid Volumetric Imaging of Cardiac Contraction
Scientists have developed a new light-sheet microscopy platform capable of capturing rapid, three-dimensional cardiac contractions with unprecedented speed and minimal light exposure. The system overcomes limitations of existing techniques by integrating compressive sensing with light-sheet microscopy, achieving volumetric imaging rates of up to 200 volumes per second while maintaining cellular resolution, delivering a significant improvement in spatiotemporal fidelity for observing dynamic cardiac processes. The core of the system lies in a compressed acquisition strategy, where fluorescence signals from multiple depth planes are encoded within a single exposure using a digital micromirror device. This device rapidly projects programmable binary patterns, spatially encoding the fluorescence, and combines it with continuous axial scanning to capture data efficiently.
By operating at a given compression ratio, the required data storage is reduced by the same factor, providing a clear advantage for high-speed volumetric imaging. To reconstruct the images from these encoded signals, researchers propose a novel reconstruction framework that addresses challenges arising from the superposition of depth layers and correlations between adjacent slices. The team employs a Plug-and-Play framework, solved using the alternating direction method of multipliers, which flexibly incorporates advanced denoisers, including Tikhonov, Total Variation, and BM3D. Furthermore, they introduced temporal regularization, enforcing smoothness between adjacent z-slices to preserve structural continuity in the dynamic imaging process. Experiments on zebrafish heart imaging demonstrate that the method successfully reconstructs cellular structures with excellent denoising performance and image clarity, validating the effectiveness and robustness of the algorithm in real-world, high-speed, low-light biological imaging scenarios.
Zebrafish Heart Imaging via Temporal Reconstruction
This research presents a new imaging framework that combines compressive sensing with light-sheet microscopy, enabling high-performance, low-phototoxic imaging of the beating heart. The team developed a flexible reconstruction approach using a plug-and-play algorithm, incorporating various image denoisers including Tikhonov, total variation, and BM3D. A key innovation lies in the introduction of temporal regularization, which leverages correlations between successive image slices to improve reconstruction quality and preserve structural continuity during dynamic imaging. Experimental results on zebrafish heart imaging demonstrate the effectiveness of this method, successfully reconstructing cellular structures even with high compression ratios.
The research reveals a trade-off between different image priors, with BM3D offering the highest fidelity, total variation balancing quality and computational cost, and Tikhonov providing the fastest processing speed. The framework’s adaptability and performance establish a practical basis for high-speed, low-dose volumetric microscopy, particularly suited for capturing dynamic biological processes like cardiac function. Future research will extend this approach to simultaneously reconstruct multiple time points within the cardiac cycle, ultimately enabling true 4D volumetric reconstruction that captures both the spatial and temporal dynamics of the beating heart.
👉 More information
🗞 A Plug-and-Play Framework for Volumetric Light-Sheet Image Reconstruction
🧠 ArXiv: https://arxiv.org/abs/2511.03093