Understanding how genes activate within tissues relies on identifying communities of cells that share similar characteristics, known as spatial domains, but current methods often overlook the complex interplay between structures at different scales. Perry Beamer and Zixuan Cang, from North Carolina State University, alongside their colleagues, address this challenge with a new approach called PHD-MS, which leverages the mathematical tools of topological data analysis to pinpoint tissue structures that persist across multiple morphological scales. This innovative method reveals previously hidden relationships within tissues, offering a more comprehensive understanding of gene expression patterns, and importantly, outperforms existing clustering techniques when tested against expert-annotated tissue samples. By providing a powerful, open-source software package with an interactive interface, PHD-MS empowers researchers to explore these multiscale domains and unlock new insights into tissue organisation and function.
Mapping Gene Expression Within Tissue Context
Spatial transcriptomics is a rapidly evolving field focused on measuring gene expression while simultaneously preserving information about the location of genes within a tissue, allowing researchers to understand how gene activity relates to the tissue’s overall structure and organisation. Several technologies are now available to generate spatial transcriptomic data, including methods that enable the measurement of many genes across a tissue section and techniques like osmFISH, which visualise and quantify RNA molecules with high resolution. A new unsupervised method, SCALE, identifies multi-scale domains within spatial omics data, revealing patterns at different levels of granularity. Analysing spatial transcriptomic data requires specialised computational tools, such as community detection algorithms like Louvain and Leiden, which identify clusters of cells based on their gene expression patterns. Persistent homology, a technique from topological data analysis, identifies and characterises the shape of data, revealing underlying structures and relationships, while optimal transport provides a mathematical framework for comparing gene expression patterns across different spatial locations. These methods are applied to a wide range of biological contexts, including cancer research and brain research, providing insights into disease mechanisms and neural circuit organisation.
Multiscale Tissue Structure via Persistent Homology
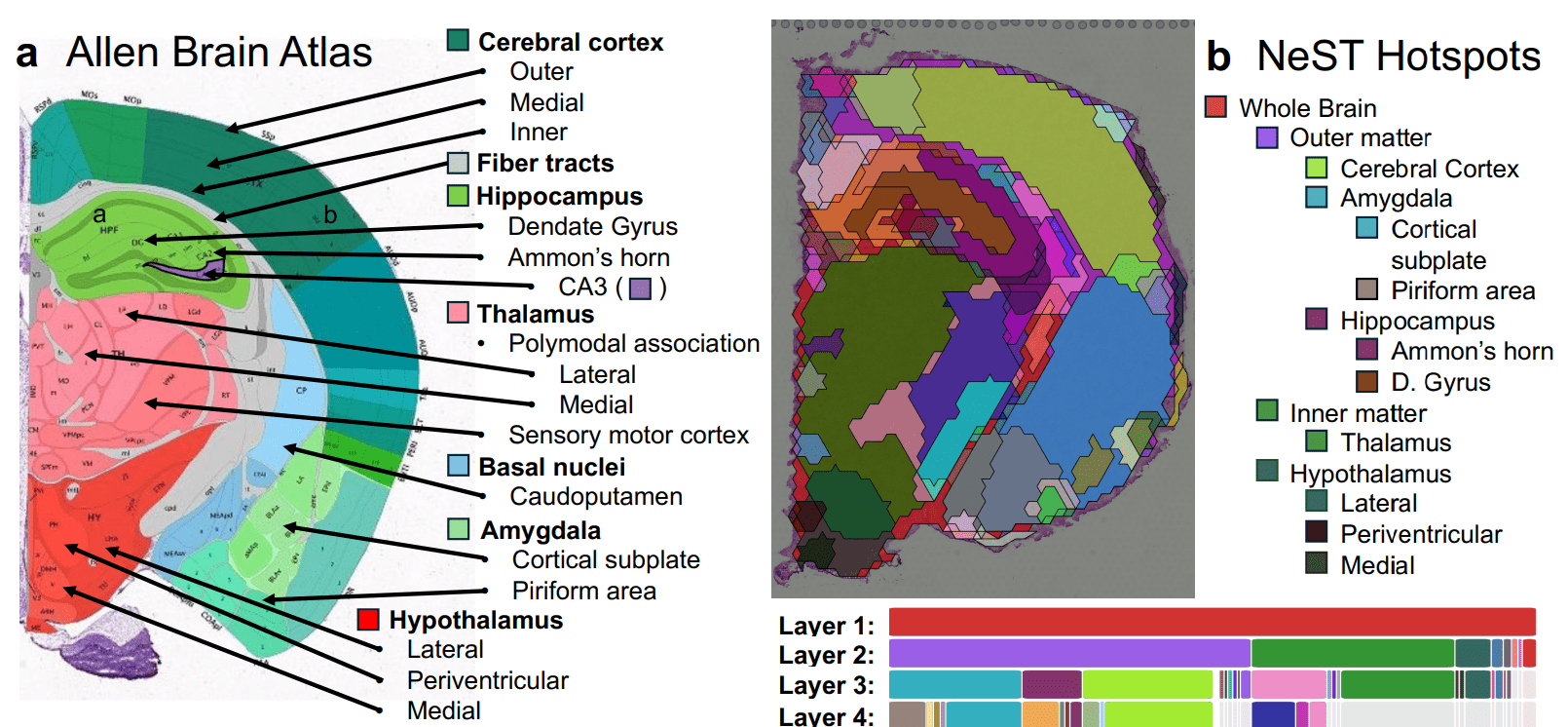
Researchers have developed Persistent Homology for Domains at Multiple Scales (PHD-MS), a new computational approach for analysing spatial transcriptomics data and identifying tissue structures across varying scales. Existing methods often struggle to capture the hierarchical and heterogeneous nature of biological tissues, focusing on a single scale. PHD-MS utilises topological data analysis to locate and characterise multiscale spatial domains within tissue samples, beginning with spatial transcriptomics data that maps gene expression to precise locations. Researchers apply persistent homology to these data, identifying connected components and holes that persist across a range of scales, revealing stable tissue structures regardless of the chosen resolution. PHD-MS identifies nested structures, mirroring the biological organisation of tissues like the hippocampus and its subfields, and demonstrates superior performance compared to traditional clustering approaches when compared against expert-annotated tissues. The team has released PHD-MS as open-source software with an interactive graphical user interface, facilitating exploration of the identified multiscale domains and providing a systematic approach to summarise stable structures across scales.
Multiscale Tissue Structures Revealed by PHD-MS
Scientists have developed a novel approach to spatial transcriptomics data analysis, introducing Persistent Homology for Domains at Multiple Scales (PHD-MS) to identify tissue structures that persist across varying scales. This work addresses the limitations of existing methods, which typically overlook the naturally multiscale and heterogeneous nature of biological tissues. The team constructed a spatial cluster filtration, enabling connections between tissue domains to be identified at multiple spatial resolutions, and then applied persistent homology to reveal prominent multiscale patterns. Experiments demonstrate that PHD-MS effectively locates domains that remain stable across multiple resolutions and identifies interactions between domains by assessing their similarity across scales.
In mouse brain data, PHD-MS successfully resolved the hierarchical organisation of subregions, highlighting nested structures within the tissue. Applying PHD-MS to breast cancer data distinguished stable and unstable regions of the tumour microenvironment, accurately corresponding to mixed border regions between malignant and healthy tissue, and integrated differential gene expression data, revealing scale-dependent transcriptional patterns within tumours. Quantitative benchmarking consistently showed that PHD-MS improves domain identification compared to single-scale analyses, and the team implemented a point-and-click visualisation utility, allowing researchers to easily explore PHD-MS domains around specific points within a tissue sample.
Multiscale Tissue Domains via Persistent Homology
This research presents a new computational method, Persistent Homology for Domains at Multiple Scales (PHD-MS), designed to improve the analysis of spatial transcriptomics data. Existing methods for identifying spatial domains often focus on a single scale of tissue organisation, potentially overlooking important relationships between large structures and finer substructures within them. PHD-MS utilises techniques from topological data analysis to detect and characterise these multiscale domains, effectively bridging gaps between different levels of tissue organisation. The team demonstrated PHD-MS’s effectiveness by applying it to several tissue types and spatial transcriptomics technologies, and the method outperformed traditional clustering approaches when compared against expert-annotated tissues, indicating its ability to accurately identify biologically relevant structures. The researchers acknowledge that the performance of PHD-MS, like other methods in this field, is dependent on the quality and resolution of the input spatial transcriptomics data, and have made the software openly available, complete with a user-friendly interface, to facilitate further exploration and application of this approach by the wider research community. This work provides a valuable new tool for investigating tissue organisation and understanding the complex interplay of gene expression and spatial context.
👉 More information
🗞 PHD-MS: Multiscale Domain Identification for Spatial Transcriptomics via Persistent Homology
🧠 ArXiv: https://arxiv.org/abs/2511.08411