Researchers have developed La-Proteina, a new framework capable of generating diverse and co-designable protein structures at an unprecedented scale, achieving state-of-the-art performance in fully atomistic protein design. The system generates proteins with backbones of up to 800 residues, surpassing previous methods, and demonstrates superior structural validity as assessed by MolProbity. La-Proteina also excels in fully atomistic motif scaffolding, completing tasks where a comparable system, Protpardelle, largely fails, indicating potential for applications in enzyme design and binder development.
Novel Protein Generation Framework
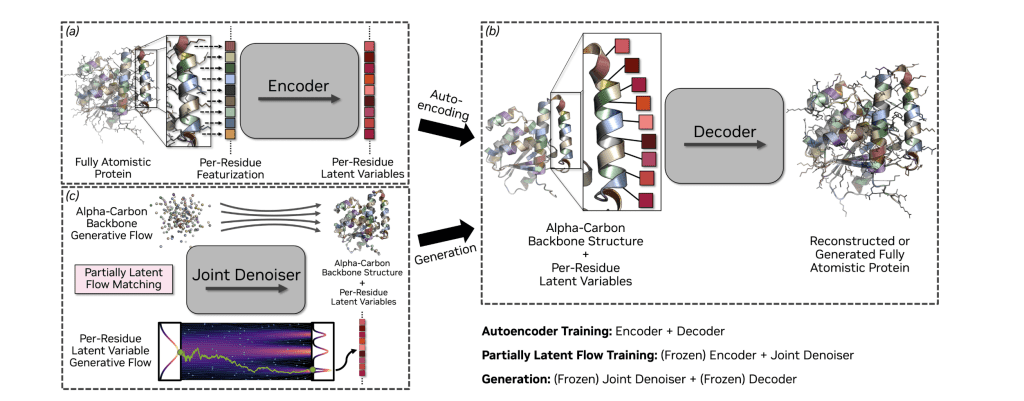
La-Proteina introduces a novel framework for atomistic protein design, based on a partially latent protein representation. This approach models coarse backbone structure explicitly, while capturing sequence and atomistic details via per-residue latent variables. By circumventing the challenges associated with explicit side-chain representations during generation, the system enables the joint generation of protein sequence and fully atomistic structure. La-Proteina employs flow matching in this partially latent space to model the joint distribution over sequences and full-atom structures.
Evaluations demonstrate that La-Proteina achieves state-of-the-art performance on multiple generation benchmarks, including all-atom designability and diversity, and is scalable to long chain generation, producing proteins of up to 800 residues. This represents a significant advancement, as La-Proteina is the first method capable of generating diverse, co-designable proteins at such lengths, despite some degradation in co-designability at longer lengths. The efficiency of this performance is attributed to the system’s partially latent flow matching framework.
Generated structures exhibit superior structural validity compared to existing all-atom generation baselines, as evidenced by MolProbity assessments which demonstrate increased physical realism. Furthermore, La-Proteina accurately recovers side chain dihedral angle distributions, aligning with reference data from the Protein Databank (PDB) and AlphaFold Database (AFDB). The framework also excels in fully atomistic motif scaffolding, successfully completing functionally relevant motifs with fully atomistic scaffolds in both indexed and unindexed generation scenarios.
Performance and Scalability
La-Proteina demonstrates scalability to long chain generation, producing diverse and (co-)designable protein backbones of up to 800 residues, significantly outperforming previous methods. While co-designability degrades at longer lengths, La-Proteina is the first method capable of generating diverse co-designable proteins at such lengths. This capability is attributed to the system’s highly efficient partially latent flow matching framework.
Generated structures exhibit superior structural validity compared to existing all-atom generation baselines, as demonstrated by MolProbity assessments which indicate increased physical realism. La-Proteina accurately recovers side chain dihedral angle distributions, aligning with reference data from the Protein Databank (PDB) and AlphaFold Database (AFDB).
In fully atomistic motif scaffolding, La-Proteina successfully completed tasks in both indexed and unindexed generation scenarios, generating many diverse unique successes, whereas Protpardelle failed on most tasks. This highlights La-Proteina’s performance in completing functionally relevant motifs with fully atomistic scaffolds.
Structural Validity and Applications
La-Proteina produces structures with higher structural validity than existing all-atom generation baselines, as demonstrated by MolProbity assessments which demonstrate increased physical realism. Furthermore, the system accurately recovers side chain dihedral angle distributions, aligning with reference data from the Protein Databank (PDB) and AlphaFold Database (AFDB).
La-Proteina excels in fully atomistic motif scaffolding, where a functionally relevant motif is completed with a fully atomistic scaffold, tackling both all-atom and tip-atom scaffolding in indexed and unindexed generation scenarios. In these tasks, La-Proteina generated many diverse unique successes, whereas Protpardelle failed on most tasks.
The system’s performance highlights its relevance for important conditional atomistic protein generation tasks, such as enzyme design, and demonstrates potential for unlocking large-scale fully atomistic protein structure generation. Future work could apply La-Proteina to challenging binder design problems.
More information
External Link: Click Here For More




