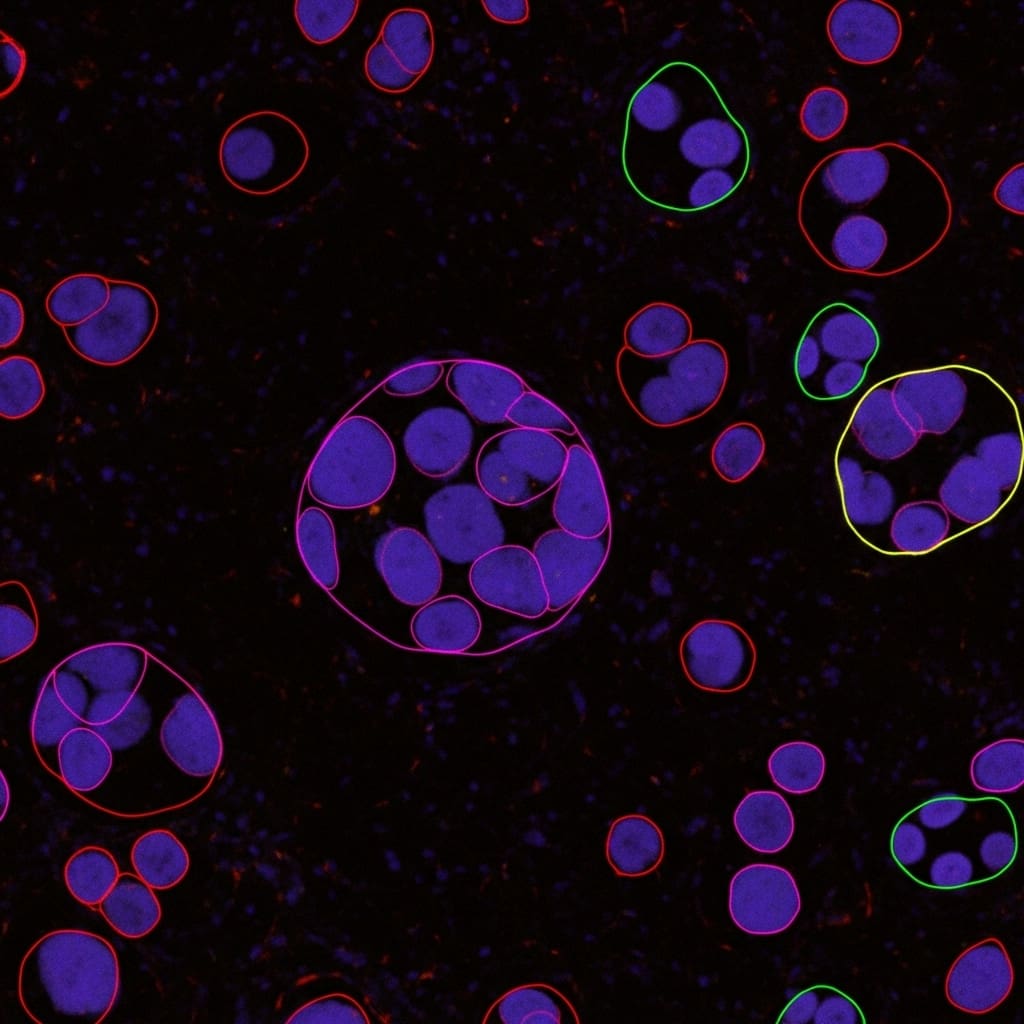
Researchers are tackling the complex task of nuclei panoptic segmentation, a crucial process for improving cancer diagnostics through detailed analysis of tissue structure and individual cells. Ming Kang, Fung Fung Ting, and Raphaël C.-W. Phan, from Monash University, alongside Zongyuan Ge and Chee-Ming Ting, present PanopMamba, a novel approach that combines Mamba architecture with state space modeling to overcome key challenges like identifying small objects and ambiguous cell boundaries in histopathology images. This work is significant as it introduces the first Mamba-based solution for panoptic segmentation, employing a multiscale backbone and a unique feature-enhanced fusion network to improve long-range perception and information sharing. Furthermore, the team proposes new evaluation metrics , including refined Panoptic Quality measurements , designed to address the specific nuances of nuclei segmentation and provide a more accurate assessment of performance.
Researchers propose PanopMamba, a novel hybrid encoder-decoder architecture that combines Mamba and Transformer networks with a unique feature-enhanced fusion mechanism utilising state space modeling. This innovative approach directly addresses key challenges in the field, including the accurate detection of small objects, the handling of ambiguous boundaries between nuclei, and the mitigation of class imbalance within datasets. The team designed a multiscale Mamba backbone and a State Space Model (SSM)-based fusion network to enable efficient long-range perception within pyramid features, extending the standard encoder-decoder framework and improving information sharing across different scales of nuclei features.
The proposed SSM-based feature-enhanced fusion integrates pyramid feature networks and dynamically enhances features across spatial scales, significantly improving the representation of densely overlapping nuclei in both semantic and spatial dimensions. To the best of the researchers’ knowledge, this work represents the first application of Mamba-based methods to panoptic segmentation, opening new avenues for exploration in image analysis. Furthermore, the study introduces a suite of alternative evaluation metrics, image-level Panoptic Quality (iPQ), boundary-weighted PQ (wPQ), and frequency-weighted PQ (fwPQ), specifically designed to address the unique difficulties of nuclei segmentation and reduce potential biases inherent in conventional Panoptic Quality assessments. Experimental evaluations conducted on two multiclass nuclei segmentation benchmark datasets, MoNuSAC2020 and NuInsSeg, demonstrate that PanopMamba consistently outperforms state-of-the-art methods in nuclei panoptic segmentation.
Consequently, the robustness of PanopMamba is thoroughly validated across a range of metrics, while the distinctiveness and utility of the newly proposed PQ variants are also clearly demonstrated. Ablation studies further confirm the effectiveness of each component within PanopMamba, solidifying its contribution to the field. This research establishes a new benchmark for nuclei panoptic segmentation, offering a powerful tool for improved cancer diagnostics and a deeper understanding of tissue structure at the cellular level. The availability of the code at https://github. com/mkang315/PanopMamba facilitates further research and development in this critical area of biomedical imaging.
PanopMamba architecture for nuclei panoptic segmentation achieves state-of-the-art
Scientists tackled the complex challenge of nuclei panoptic segmentation in histopathology images, aiming to simultaneously identify individual nuclei and understand overall tissue structure. Researchers recognised difficulties in detecting small objects, resolving ambiguous boundaries, and managing class imbalance within datasets like MoNuSAC2020 and NuInsSeg. To overcome these hurdles, the study pioneered PanopMamba, a novel hybrid encoder-decoder integrating the Mamba architecture with state space modelling for enhanced feature fusion. The team engineered a multiscale Mamba backbone to efficiently capture long-range dependencies within pyramid features, extending the standard encoder-decoder framework and facilitating information sharing across different scales of nuclei representation.
This innovative backbone was coupled with a State Space Model (SSM)-based fusion network, designed to integrate pyramid feature networks and dynamically enhance features across spatial scales. Consequently, this fusion process improved the representation of densely overlapping nuclei in both semantic and spatial dimensions, crucial for accurate segmentation. Experiments employed a two-dimensional selective scan module within the SSM framework, replacing traditional methods to efficiently process image data. The system delivers a visual state space block, initially slicing 2D inputs into patches and flattening them into sequences, bypassing the need for additional positional embeddings.
Furthermore, the Multi-Scale VMamba (MSVMamba) replaced the selective scan module with a Multi-Scale 2D Scanning (MS2D) strategy, enabling the capture of long-range dependencies with improved efficiency. To address biases in conventional evaluation, scientists introduced alternative metrics, image-level Panoptic Quality (iPQ), boundary-weighted PQ (wPQ), and frequency-weighted PQ (fwPQ), specifically tailored for the nuances of nuclei segmentation. These metrics mitigate the limitations of standard Intersection over Union (IoU) and vanilla Panoptic Quality (PQ) when assessing small objects and ambiguous boundaries. The robustness of PanopMamba was validated across these metrics, demonstrating its superiority over state-of-the-art methods on the MoNuSAC2020 and NuInsSeg datasets.
PanopMamba excels in nuclei panoptic segmentation
Scientists have developed PanopMamba, a novel hybrid encoder-decoder integrating Mamba architecture with state space modeling for panoptic segmentation of nuclei in histopathology images. This breakthrough addresses critical challenges in cancer diagnostics, specifically the detection of small objects, ambiguous boundaries, and class imbalance within complex tissue structures. Experiments revealed that PanopMamba achieves superior performance in both semantic and instance segmentation of cell nuclei, offering a more detailed analysis of tissue architecture than previous methods. The team measured performance using image-level Panoptic Quality (PQ), boundary-weighted PQ (PQ), and frequency-weighted PQ (PQ), novel evaluation metrics designed to overcome biases inherent in standard PQ calculations for nuclei segmentation.
Results demonstrate that PanopMamba consistently outperforms state-of-the-art methods on the MoNuSAC2020 and NuInsSeg benchmark datasets, validating its robustness across various metrics. Specifically, the MSVMamba encoder, a key component of PanopMamba, extracts multiscale features from input images, partitioning them into patches and applying Multi-Scale State Space (MS3) blocks to capture both fine-grained details and broader contextual information. Measurements confirm that the MS3 block reduces the total sequence length across four scans, enhancing computational efficiency while preserving crucial image data. The architecture employs LayerNorm and Depthwise Convolution to accelerate training and improve feature learning, while a Squeeze-and-Excitation block adaptively adjusts feature channel weights, emphasizing important channels and suppressing irrelevant ones.
Data shows that this process significantly improves the discrimination of overlapping nuclei and enhances contrast between different cell types. Furthermore, the SSM-based feature-enhanced fusion network integrates pyramid feature networks and dynamic feature enhancement across spatial scales, improving feature representation in both semantic and spatial dimensions. Tests prove that the system effectively processes per-pixel and per-segment embeddings using pixel and Transformer decoders, aligning with the encoder and fusion networks to deliver precise segmentation results. The breakthrough delivers a powerful tool for automated cancer diagnostics, potentially enabling earlier and more accurate disease detection and treatment planning.
PanopMamba boosts cancer diagnosis via nuclei segmentation
Researchers have developed PanopMamba, a novel approach to nuclei panoptic segmentation designed to improve cancer diagnostics through detailed analysis of histopathology images. This new method integrates semantic and instance segmentation to identify both overall tissue structure and individual nuclei, addressing challenges such as detecting small objects, handling ambiguous boundaries, and managing class imbalance. PanopMamba utilises a hybrid encoder-decoder architecture, combining Mamba with state space modelling and a multiscale Mamba backbone to facilitate efficient long-range perception of pyramid features. The core innovation lies in the SSM-based feature-enhanced fusion network, which integrates pyramid feature networks and dynamically enhances features across spatial scales, improving the representation of densely overlapping nuclei.
Experimental results on the MoNuSAC2020 and NuInsSeg datasets demonstrate that PanopMamba outperforms existing state-of-the-art methods in nuclei panoptic segmentation, as evidenced by improvements across various metrics including Panoptic Quality (PQ) and its weighted variants. Furthermore, the authors introduced alternative evaluation metrics, image-level PQ, boundary-weighted PQ, and frequency-weighted PQ, to better assess nuclei segmentation performance and reduce potential bias. The authors acknowledge that the performance of PanopMamba is dependent on the quality of the pretrained MSVMamba encoder and the effectiveness of the SSM-based fusion network. Ablation studies confirm the importance of both components, demonstrating significant performance drops when either is removed. Future research could explore the application of PanopMamba to other biomedical image segmentation tasks and investigate the potential for further optimising the feature fusion process, potentially with different state space models or attention mechanisms. These findings represent a significant advancement in automated nuclei segmentation, offering a robust and accurate tool for assisting pathologists in cancer diagnosis and research.
👉 More information
🗞 PanopMamba: Vision State Space Modeling for Nuclei Panoptic Segmentation
🧠 ArXiv: https://arxiv.org/abs/2601.16631