Researchers developed new methods to quantify the probability of specific tree topologies within Bayesian phylogenetic analyses. Algorithms efficiently estimate ‘credible clade distributions’, representing sets of trees collectively accounting for a defined probability. Validation using simulations and real datasets demonstrates improved accuracy and facilitates robust model evaluation beyond standard coverage analyses.
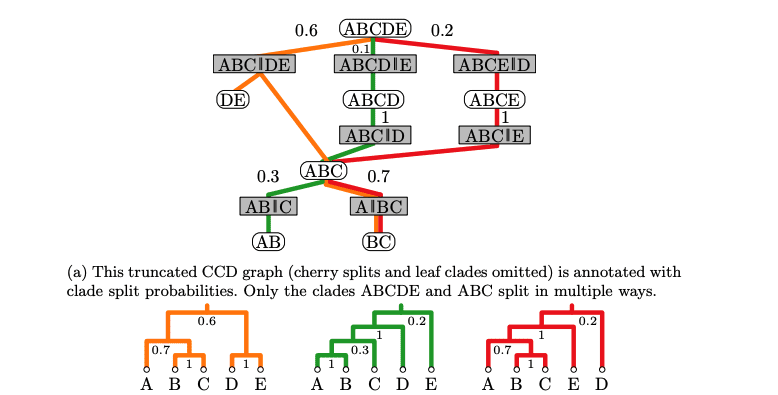
Understanding evolutionary relationships requires constructing phylogenetic trees – branching diagrams depicting the inferred ancestry of species. However, representing the uncertainty inherent in these reconstructions presents a statistical challenge, particularly when considering the immense number of possible tree topologies. Researchers are now refining methods to quantify the credibility of these topologies, moving beyond simple frequency-based approaches which struggle with complex datasets. Jonathan Klawitter and Alexei J. Drummond, both from the University of Auckland, alongside colleagues, detail these advances in their paper, “Credible Sets of Phylogenetic Tree Topology Distributions”, introducing Conditional Clade Distributions (CCDs) – statistical models representing the probability of specific groupings of species – and a novel concept termed ‘credible CCDs’ to more accurately define the range of plausible evolutionary trees.
Defining Phylogenetic Relationships: Improved Methods for Credible Sets
Traditional methods for representing uncertainty in evolutionary relationships often struggle with complex datasets, prompting the development of more robust approaches for phylogenetic inference. This research introduces algorithms utilising Conditional Clade Distributions (CCDs) – a statistical approach modelling the distribution of possible evolutionary trees – to more accurately define credible sets of phylogenetic trees, offering a framework for assessing evolutionary uncertainty and enabling more informed conclusions about evolutionary history.
Evaluations utilising simulated datasets – including Yule50, DS3-4, and DS1-2 – demonstrate the performance of these methods in both accuracy and stability. Accuracy, in this context, refers to the ability to include the true evolutionary tree within the credible set, while stability reflects the consistency of results across different analyses and parameter settings. The algorithms were rigorously tested under various conditions, ensuring their reliability in handling complex phylogenetic problems. Results indicate that the choice of method significantly impacts both sensitivity – the ability to correctly include trees that should be within the set – and specificity – the ability to exclude those that should not, demonstrating the importance of selecting an appropriate analytical framework.
The study highlights the critical influence of sample size on the stability of probability-based credible sets, emphasising the need for sufficient computational resources and careful consideration of sampling strategies. Analyses of the Yule50 dataset reveal that increasing the sample size reduces variance in the computed credible level, suggesting a sample size of 10,000 trees provides acceptable stability and ensures the reliability of the inferred phylogenetic relationships. Researchers carefully examined the trade-off between computational cost and analytical precision, identifying an optimal balance that maximises the information gained from the analysis.
Beyond defining credible sets, the authors validate their methods through rank-uniformity validation and the generation of Empirical Cumulative Distribution Function (ECDF) plots, providing a comprehensive assessment of model performance and uncertainty. Rank-uniformity validation assesses the method’s ability to correctly rank the probability of different trees, while ECDF plots visualise the distribution of probabilities, providing insights into the uncertainty associated with the inferred phylogeny.
Future work will extend these methods to accommodate larger and more complex datasets, including those derived from genomic-scale data, pushing the boundaries of phylogenetic inference. Investigating the impact of different CCD models on the accuracy and efficiency of credible set construction also warrants further attention, allowing for the optimisation of the analytical framework. Additionally, exploring the application of these methods to other areas of phylogenetic inference, such as ancestral state reconstruction and divergence time estimation, could broaden their impact and utility. Researchers plan to collaborate with experts in computational biology and statistics to develop innovative algorithms and analytical tools that address the challenges of large-scale phylogenetic inference.
This research introduces a novel approach to phylogenetic inference, offering an improvement over traditional methods and providing a more robust framework for understanding evolutionary relationships. By leveraging the power of Conditional Clade Distributions and employing rigorous validation techniques, researchers have developed a tool that will advance the field of evolutionary biology. The findings of this study have implications for a range of applications, including conservation biology, disease epidemiology, and comparative genomics.
👉 More information
🗞 Credible Sets of Phylogenetic Tree Topology Distributions
🧠 DOI: https://doi.org/10.48550/arXiv.2505.14532