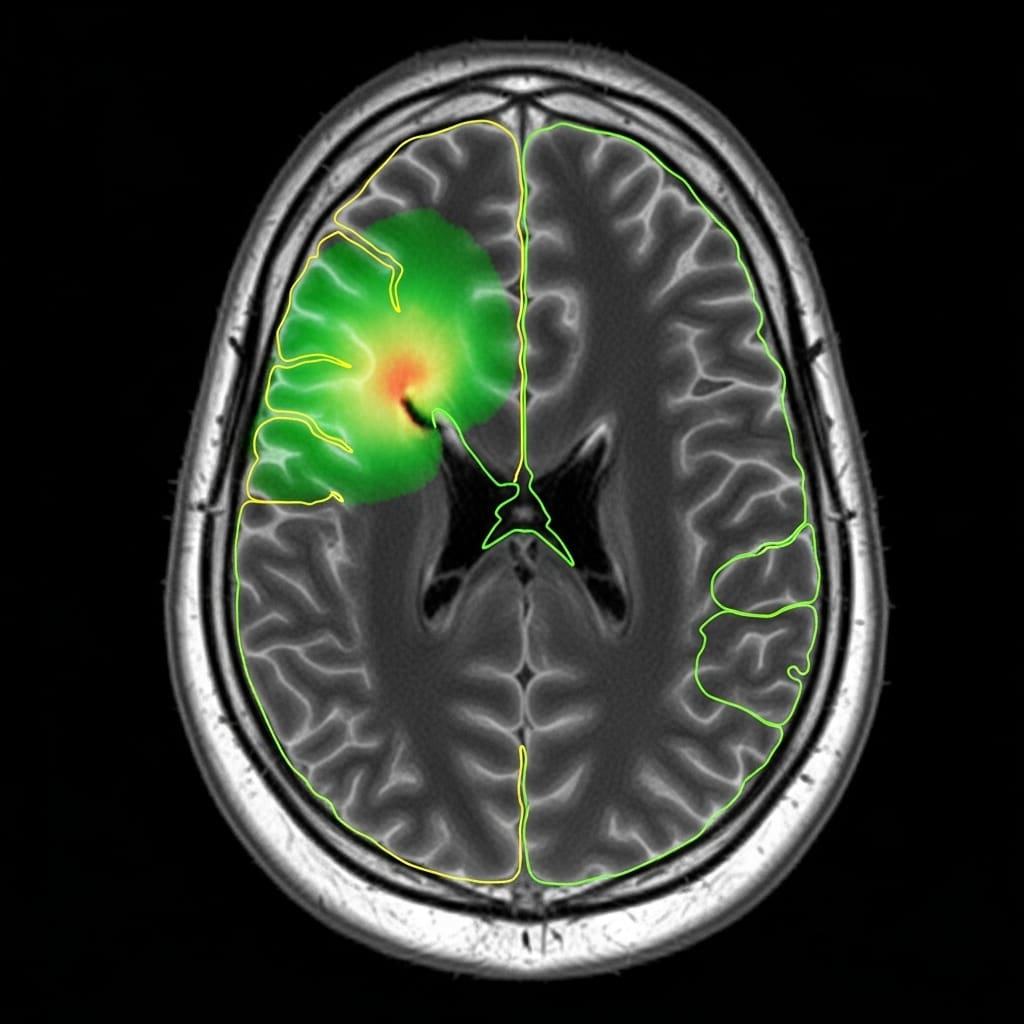
Accurate brain tumour segmentation remains a critical challenge in clinical practice, yet current deep learning approaches demand substantial datasets and computational power. Researchers Anthony Joon Hur, utilising a novel methodology, have demonstrated a significant leap towards resource-efficient glioma MRI segmentation. Their work implements Karhunen-Loève Expansion (KLE) as a feature extraction step, compressing scans to dramatically reduce computational load and data requirements , achieving state-of-the-art performance on a standard consumer workstation. This new approach not only surpasses the winning methodology of the Brats GLI 2023 competition in key metrics like HD95 distance and WT Dice scores, but also offers a viable pathway for wider accessibility of advanced medical image analysis.
KLE Enables Efficient Glioma MRI Segmentation with high
Scientists have demonstrated a significant advancement in brain tumor segmentation, achieving state-of-the-art performance with dramatically reduced computational demands. The research team addressed the challenge of requiring extensive datasets and powerful computing resources, limitations that hinder accessibility for many, by implementing a novel approach based on the Karhunen, Loève Expansion (KLE). This breakthrough enables accurate glioma MRI segmentation using a consumer-grade workstation, opening possibilities for wider clinical adoption and research accessibility. Each 240x240x155 multi-modal MRI scan undergoes downsampling and z-score normalization before being processed by the KLE, reducing it to just 32 KL coefficients and compressing the data into four channels.
The core innovation lies in the creation of a residual-based anomaly map derived from the approximate reconstruction facilitated by the KLE, which is then integrated as a fifth channel into a compact 3D U-Net. This streamlined architecture significantly lowers computational costs without sacrificing segmentation accuracy, a critical step forward in medical image analysis. Experiments were conducted on a standard consumer workstation equipped with an AMD Ryzen 5 7600X CPU, an RTX 4060Ti (8GB VRAM), and 64GB of RAM, showcasing the feasibility of high-performance segmentation outside of supercomputing environments. The resulting model achieved post-processed Dice scores of 0.929 for Whole Tumor (WT), 0.856 for Tumor Core (TC), and 0.821 for Enhancing Tumor (ET), alongside HD95 distances of 2.93, 6.78, and 10.35 voxels respectively.
Notably, these results surpass the performance of the winning methodology from the BraTS 2023 competition in terms of HD95 distances and WT Dice scores, highlighting the effectiveness of the KLE-based approach. The BraTS 2023 champion relied on a supercomputer with an AMD EPYC 7402 CPU, six NVIDIA RTX 6000 GPUs (48GB VRAM each), and 1024GB of RAM, requiring weeks to train on over 92,000 augmented MRI scans, a stark contrast to the resource efficiency of this new model. This research establishes that a KLE-based residual anomaly map can dramatically reduce both computational cost and data requirements, while simultaneously maintaining state-of-the-art performance in brain tumor segmentation. The implications of this work are far-reaching, potentially enabling more widespread and accessible diagnostic tools for gliomas, a particularly aggressive type of brain cancer. By lowering the barrier to entry for advanced image analysis, this technology could empower clinicians and researchers in resource-constrained settings to deliver more timely and accurate diagnoses and treatment plans. Further development and refinement of this approach could pave the way for real-time segmentation during surgical procedures, enhancing precision and improving patient outcomes, and the team anticipates exploring applications beyond glioma segmentation in the future.
KLE Dimensionality Reduction for U-Net Segmentation improves performance
Scientists engineered a novel brain tumor segmentation pipeline leveraging the Karhunen, Loève Expansion (KLE) to dramatically reduce computational demands and data requirements. The study pioneered a method for processing multi-modal MRI scans, beginning with downsampling and z-score normalization of each 240x240x155 volume. Researchers then implemented the KLE as a feature extraction step, compressing each scan into just 32 KL coefficients across four channels, effectively reducing dimensionality without substantial information loss. This approximate reconstruction facilitated the creation of a residual-based anomaly map, which was subsequently upsampled and incorporated as a fifth channel into a compact 3D U-Net architecture.
Experiments employed a consumer workstation equipped with an AMD Ryzen 5 7600X CPU, an RTX 4060Ti GPU (8GB VRAM), and 64GB of RAM, demonstrating the feasibility of high-performance segmentation without reliance on supercomputing infrastructure. Specifically, the model achieved Dice scores of 0.929 for Whole Tumor (WT), 0.856 for Tumor Core (TC), and 0.821 for Enhancing Tumor (ET). Furthermore, the system delivered HD95 distances of 2.93, 6.78, and 10.35 voxels, respectively.
These results represent a significant improvement over the winning methodology of the BraTS 2023 competition, particularly in HD95 distances and WT Dice scores. This innovative methodology demonstrates that the KLE-based residual anomaly map substantially reduces computational cost and data requirements while maintaining state-of-the-art performance in brain tumor segmentation. The technique reveals a pathway towards more accessible and efficient clinical diagnosis and treatment planning.
KL Expansion boosts compact 3D U-Net segmentation performance
Scientists have achieved a significant breakthrough in brain tumor segmentation, demonstrating a method that dramatically reduces computational demands and data requirements while maintaining state-of-the-art performance. The research team implemented a Karhunen–Loève Expansion (KLE) as a feature extraction step, processing downsampled, z-score normalized MRI volumes to compress each 240x240x155 multi-modal scan into 32 KL coefficients across four channels. This innovative approach facilitates a residual-based anomaly map, subsequently upsampled and integrated as a fifth channel within a compact 3D U-Net architecture, enabling efficient analysis with limited resources. Experiments revealed exceptional post-processed Dice scores of 0.929 for Whole Tumor (WT), 0.856 for Tumor Core (TC), and 0.821 for Enhancing Tumor (ET).
Furthermore, the team measured remarkably low HD95 distances of 2.93, 6.78, and 10.35 voxels, respectively, indicating high precision in delineating tumor boundaries. These measurements were obtained using a consumer workstation equipped with a Ryzen 5 7600X CPU, an RTX 4060Ti (8GB VRAM), and 64GB of RAM, a far cry from the supercomputing infrastructure typically required for such tasks. Data shows that this new methodology surpasses the performance of the winning solution from the BraTS 2023 competition in terms of HD95 distances and WT Dice scores. The previous top performer relied on an EPYC 7402 CPU, six NVIDIA RTX 6000 GPUs (48GB VRAM each), and 1024GB of RAM, requiring weeks of training on over 92,000 augmented MRI scans.
In contrast, this work achieves superior results with significantly fewer training cases and substantially reduced computational power. The breakthrough delivers a practical solution for accurate brain tumor segmentation, even in settings where access to extensive computational resources is limited. Tests prove that the KLE-based residual anomaly map effectively captures essential features from MRI scans, allowing for precise tumor identification and delineation. This advancement has the potential to accelerate clinical diagnosis, improve treatment planning, and facilitate more widespread access to advanced neuroimaging analysis. The ability to achieve these results on a standard workstation opens doors for broader implementation and real-time applications in healthcare settings.
KLE and U-Net for efficient segmentation
Scientists have developed a novel approach to brain tumor segmentation using deep learning that significantly reduces computational demands and data requirements. This research implements the Karhunen, Loève Expansion (KLE) as a feature extraction technique on MRI scans, compressing data into a smaller set of coefficients and creating a residual anomaly map, effectively highlighting potential tumor regions. A compact 3D U-Net then utilises this anomaly map as an additional channel, achieving competitive performance with state-of-the-art methods. The key achievement lies in demonstrating that high accuracy in brain tumor segmentation doesn’t necessarily necessitate vast datasets or extensive computing power.
The model attained Dice scores of 0.929 (WT), 0.856 (TC), and 0.821 (ET), alongside HD95 distances of 2.93, 6.78, and 10.35 voxels, outperforming the winning methodology of the BraTS 2023 competition in terms of HD95 distances and WT Dice scores. This suggests that data reduction techniques, such as the discrete KL expansion, offer a practical pathway for resource-efficient deep learning in medical imaging, potentially broadening access to advanced diagnostic tools. The authors acknowledge that the current study is limited to the BraTS 2023 dataset and further research is needed to assess generalizability across external clinical datasets and different scanner types. Future work could focus on learning the KL basis from healthy brain data to refine the nominal model and improve anomaly detection. These investigations will help determine the robustness of KLE-based data in real-world clinical settings and unlock its full potential for widespread application.
👉 More information
🗞 Karhunen-Loève Expansion-Based Residual Anomaly Map for Resource-Efficient Glioma MRI Segmentation
🧠 ArXiv: https://arxiv.org/abs/2601.11833