Intrinsically disordered proteins (IDPs) are increasingly recognised as vital components in many cellular processes, yet accurately modelling their behaviour remains a significant challenge for scientists. Rohan S. Adhikari, Winnie H. Shi, Amanda B. Marciel, and Walter G. Chapman, all from Rice University, investigate the performance of current protein force fields in simulating these dynamic molecules. Their work focuses on whether refinements to these force fields, combined with improved water models, can accurately predict the diverse conformations IDPs adopt in solution. The team demonstrates that the AMBER ff19SB force field, paired with the OPC water model, provides a generalised approach capable of modelling both ordered and disordered protein sequences, aligning well with experimental observations. Crucially, they also introduce a new scattering model, termed SWAXS-, which allows for detailed comparisons between simulations and experiments by accounting for the influence of hydration layers and thermal fluctuations, a particularly important consideration for understanding the behaviour of IDPs.
Molecular Dynamics Validated by X-ray Scattering
Researchers employ molecular dynamics (MD) simulations to understand how molecules behave over time, validating their accuracy using small-angle X-ray scattering (SAXS) and wide-angle X-ray scattering (WAXS) experiments. These techniques provide structural information about molecules in solution, allowing scientists to compare simulation results with real-world observations. Discrepancies between calculated and experimental profiles are minimized through data scaling and uncertainty accounting. Statistical analysis, using Chi-squared values, quantifies the goodness of fit between simulations and experiments, ensuring the reliability of the MD simulations and their ability to accurately represent molecular behavior.
Polyampholytes Validate Protein Force Field Accuracy
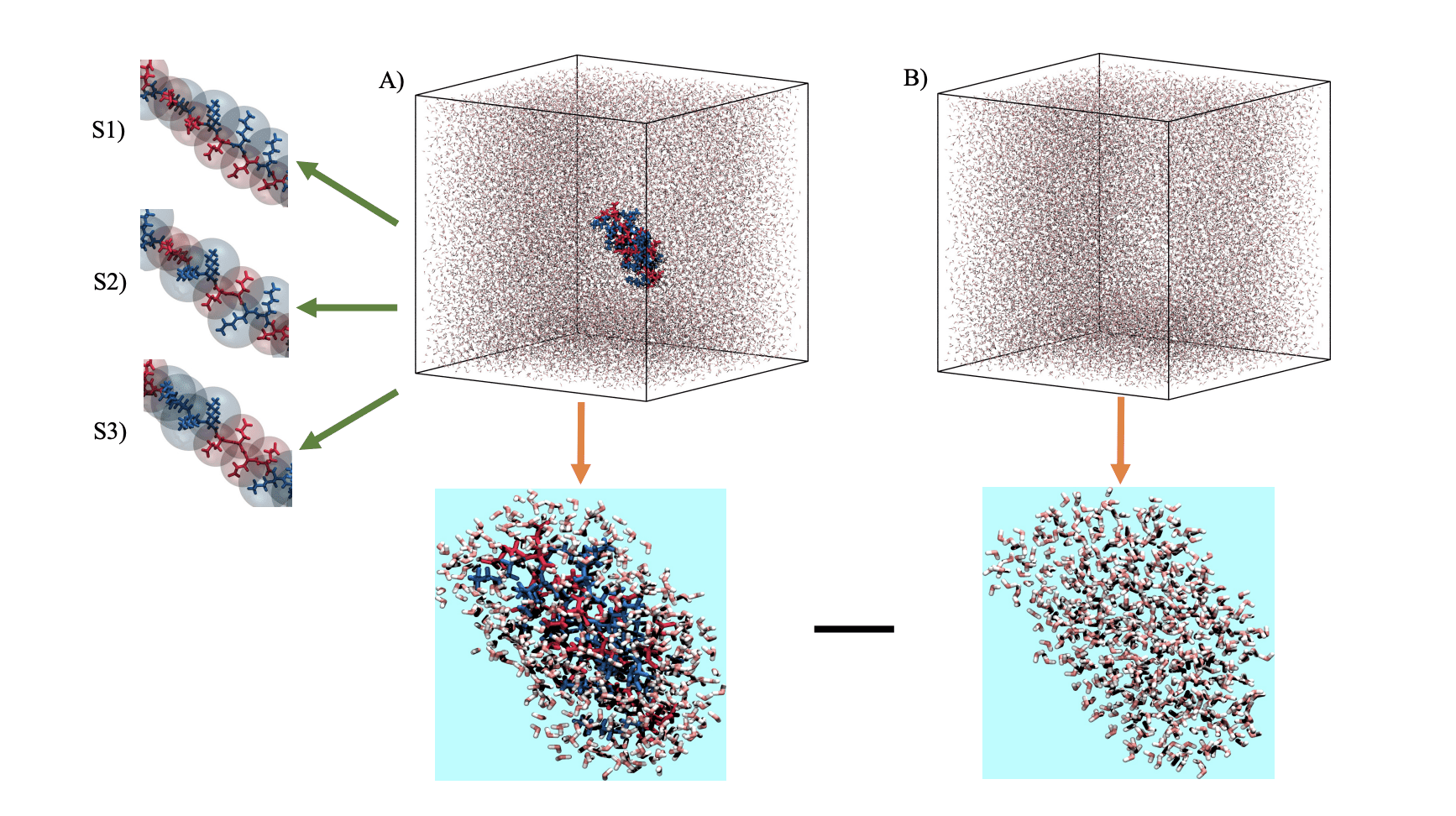
Researchers developed a comprehensive method to assess the accuracy of protein force fields and water models in simulating intrinsically disordered proteins (IDPs). Recognizing that traditional force fields are optimized for folded proteins, the team focused on polyampholytes, molecules resembling IDPs due to their simple sequences and tendency to remain disordered. The team developed a novel scattering model, termed SWAXS-, that accounts for changes in hydration layer density at the atomic level, crucial for capturing the thermal fluctuations inherent in IDPs. Traditional force fields, designed for folded proteins, often predict overly compact conformations for IDPs, hindering accurate simulations. Approximately 75% of known IDPs are polyampholytes, making these molecules ideal test systems for validating force field generalizability. By utilizing EK polyampholytes, low-complexity polymers composed of glutamic acid and lysine, researchers observed a transition from disordered to preferred conformations as the amino acid sequence was altered, mirroring the behavior of real IDPs. By applying this combination to model polyampholytes, molecules that mimic the behaviour of disordered proteins, researchers achieved close agreement between simulated and experimental small-angle X-ray scattering data, suggesting the model accurately captures the conformational diversity characteristic of these proteins in solution. The study isolated the impact of specific force field and water model improvements, confirming that the OPC water model is crucial for accurately representing disordered protein behaviour. Furthermore, the team developed a new scattering model, termed SWAXS-, which accounts for hydration layers in atomic detail, enabling more precise comparisons between simulations and experiments. While the simulations successfully reproduce experimental data, the authors acknowledge limitations in fully capturing the complexity of real biological systems and suggest future work could focus on extending this methodology to more complex IDPs and incorporating additional factors influencing protein behaviour, such as crowding and post-translational modifications.
👉 More information
🗞 The Protein Force Field Plays a Crucial Role in Obtaining Accurate Macromolecular Ensembles of IDPs
🧠 ArXiv: https://arxiv.org/abs/2508.18570