Understanding the intricate workings of individual cells requires detailed analysis of their genetic activity, but generating realistic data for testing analytical methods proves challenging. Selim Romero, Vignesh Kumar, Robert S. Chapkin, and James J. Cai from Texas A and M University present a new approach to simulating single-cell data that overcomes limitations of traditional methods. Their work introduces a quantum-enhanced simulation framework, qSimCells, which accurately models the complex, nonlinear relationships between genes and the interactions between cells by leveraging the principles of quantum entanglement. This breakthrough generates synthetic data exhibiting non-classical dependencies, revealing the inadequacy of standard analytical tools and paving the way for the development of more sophisticated techniques to decipher the complexities of gene regulation and cellular communication.
Quantum Computing Simulates Gene Regulatory Networks
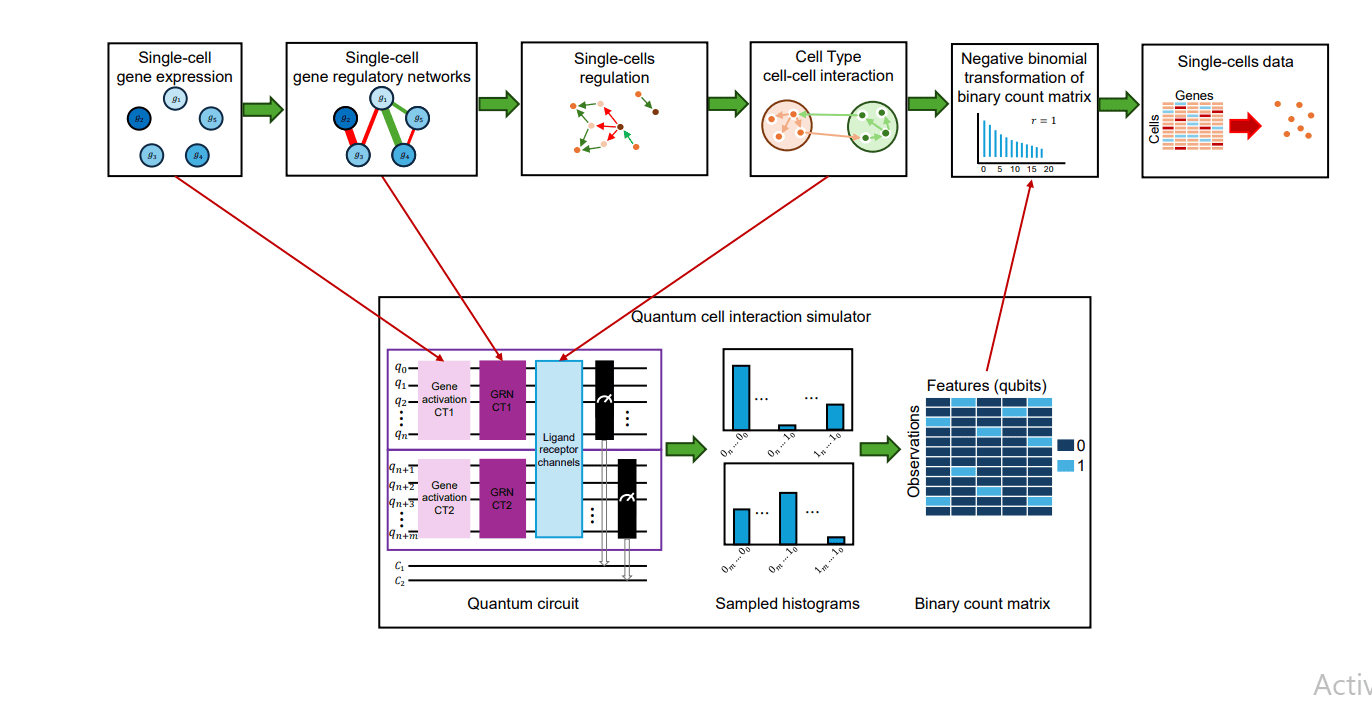
Scientists are pioneering a new approach to understanding how genes interact by using the principles of quantum computing. This research tackles the challenge of accurately modeling gene regulatory networks, which control how genes are switched on and off, from data generated by single-cell RNA sequencing. Existing methods often struggle to capture the complexity of these networks and the subtle interactions between genes, but this new technique offers a powerful alternative. The team developed qSimCells, a hybrid quantum-classical simulator that leverages quantum entanglement to model complex, non-linear dependencies within gene regulatory networks and the communication between cells.
This allows for a more realistic representation of how genes influence each other and how cells respond to signals from their environment. The method represents genes as quantum bits, or qubits, and uses carefully designed operations to simulate gene activation and interactions. The core of the method involves creating a quantum kernel, a computational structure that encodes both the internal regulatory networks within cells and the communication pathways between them. This kernel utilizes controlled-NOT (CNOT) gates to establish connections between qubits, directly modeling how one gene can influence another.
By incorporating directionality into these connections, the simulation accurately represents the causal relationships between genes, revealing which genes activate or repress others. Researchers successfully demonstrated the ability to simulate a system of ten genes, representing two different cell types, and accurately model gene activation levels. To validate the approach, the team simulated cell-cell communication and used existing analytical tools to analyze the data. Results demonstrate that standard correlation methods fail to accurately reconstruct the programmed regulatory pathways, instead reporting spurious statistical artifacts.
However, applying CellChat2. 0 to the simulated data reveals a robust increase in communication probability, up to 75-fold, when quantum entanglement is active. This confirms the essential role of the quantum kernel in creating high-fidelity data and highlights the need for advanced inference techniques to capture the complex dependencies inherent in gene regulation. This research provides a superior platform for rigorous mechanistic validation and offers a crucial step towards a more nuanced understanding of gene regulatory networks. By generating realistic data that captures the complexities of gene regulation, scientists can develop and test new analytical methods and gain deeper insights into the underlying mechanisms that control cellular behavior. Traditional simulation approaches often struggle to capture the complex, non-linear dependencies within gene regulatory networks, limiting our ability to understand how genes interact. This new technique leverages the principles of quantum computing to overcome these limitations. The core innovation lies in a quantum kernel utilizing controlled-NOT (CNOT) gates to encode both intra-cellular and inter-cellular regulatory interactions with explicit directionality, effectively modeling causality between genes.
Researchers successfully demonstrated the ability to simulate a system of ten genes, five representing each of two cell types, and accurately model gene activation levels using rotation angles that directly correspond to initial gene expression. The simulation process begins by defining an initial activation state for each gene, then applying a programmed entanglement topology to model regulatory interactions. In one demonstration, a specific inter-state cascade was established, linking genes across both cell types in a multi-step regulatory path. The resulting final quantum state was then measured multiple times to generate a histogram of measurement outcomes, forming the basis of a synthetic single-cell RNA sequencing count matrix.
To mimic the characteristics of real biological data, the researchers incorporated noise and overdispersion into the generated count matrix using a Negative Binomial distribution. Crucially, tests demonstrate that classical correlation methods, specifically Pearson and Spearman correlations, fail to accurately reconstruct the programmed regulatory paths. In contrast, applying CellChat2. 0 to the simulated data reveals a robust increase in probability, up to 75-fold, in identifying the true mechanistic links only when quantum entanglement is active. Traditional methods often struggle to capture the complex, non-linear dependencies within gene regulatory networks, hindering our understanding of how genes interact. This new technique leverages quantum entanglement to model these interactions more accurately. The team developed a kernel leveraging quantum entanglement to model complex, non-linear gene regulatory networks and cell-cell communication topologies, effectively capturing causal dependencies that classical methods struggle to represent. The simulator generates synthetic data exhibiting non-classical dependencies, providing a valuable ground truth for benchmarking advanced analytical techniques and revealing the weaknesses of traditional inference methods. Results demonstrate that standard correlation-based methods, such as Pearson and Spearman correlation, fail to accurately reconstruct programmed causal pathways.
👉 More information
🗞 A Quantum Generative Framework for Modeling Single-Cell Transcriptomes with Gene-Gene and Cell-Cell Interactions
🧠 ArXiv: https://arxiv.org/abs/2510.12776