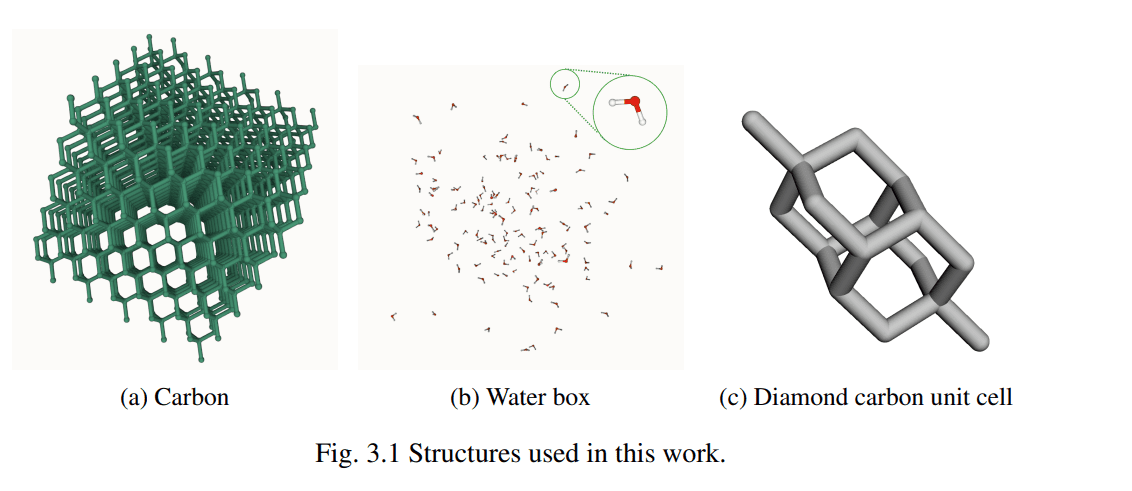
The computational cost of accurate molecular dynamics simulations often limits the size and timescale of systems scientists can study, despite advances in machine-learning force fields. Alexandre Benoit from the University of Cambridge and colleagues address this challenge by investigating methods to accelerate the MACE force field, a powerful tool for modelling molecular interactions. Their work demonstrates that employing low-precision arithmetic and optimised computational kernels can dramatically reduce simulation times without compromising the accuracy of physical results. Specifically, the team reveals that utilising the cuEquivariance backend and selectively casting linear layers to reduced precision formats delivers substantial speedups, paving the way for more efficient and larger-scale molecular simulations. This achievement promises to unlock new possibilities in fields ranging from materials science to drug discovery, where accurate and rapid simulations are essential.
This work presents original research building upon the established ACE/MACE suite of tools, adapting this architecture to explore the application of low-precision techniques. The dissertation comprises approximately 12,500 words, including appendices, bibliography, footnotes, and supporting data, with related code publicly available for reproducibility and further development. Acknowledgement is given to limitations experienced regarding access to High Performance Computing resources.
MACE Block Profiling And Bottleneck Analysis
Detailed profiling of MACE blocks, Interaction, Product, and Readout, reveals key areas for optimisation. Analysis of computational time shows that the Interaction and Product blocks are the most demanding, while the Readout block operates more quickly. The MACE/Interaction/Main and MACE/Product/Main functions consume the most processing time, followed by MACE/Interaction/SymmetricContractions and MACE/Product/SymmetricContractions. Prioritising these bottlenecks, optimisation strategies include algorithm refinement, parallelisation across multiple processor cores, and data layout improvements to maximise cache efficiency.
Specifically, the Interaction and Product blocks benefit from optimising the main functions and symmetric contractions, while the Readout block can be improved by reducing memory access and enhancing cache efficiency. Optimising linear operations using libraries like BLAS also presents opportunities for improvement, alongside careful attention to memory usage and exploring GPU acceleration with CUDA. To refine these recommendations, understanding the overall goal of the MACE implementation, the hardware configuration, the size of the input data, and the programming language and libraries used would be beneficial.
MACE Acceleration With Reduced Precision Arithmetic
This work demonstrates significant acceleration of machine-learning force fields, specifically the MACE model, while maintaining accuracy in molecular dynamics simulations. Researchers achieved speed improvements by carefully evaluating reduced-precision arithmetic and optimising GPU kernels. Profiling the MACE architecture identified opportunities for optimisation, leading to strategies that substantially reduce computational cost. Experiments show that utilising the NVIDIA cuEquivariance backend reduces inference latency by approximately three times compared to the e3nn backend. Further acceleration was achieved by casting only the linear layers to BF16 or FP16 precision within an FP32 model, yielding roughly a four-fold speedup.
Critically, these reduced-precision calculations did not compromise the accuracy of energies and thermodynamic observables in both short and long water simulations, remaining within expected run-to-run variability. Investigating the impact of different precision settings on training revealed that utilising half-precision weights degrades the accuracy of force root mean squared error, highlighting the importance of maintaining higher precision during the learning phase. Mixing the e3nn and cuEquivariance modules without explicit adapters caused representation mismatches, emphasising the need for careful integration of different computational frameworks. The research confirms that fused equivariant kernels and mixed-precision inference can significantly accelerate state-of-the-art force fields with negligible impact on downstream molecular dynamics simulations. A practical policy emerged: utilising cuEquivariance with FP32 by default, and enabling BF16/FP16 for linear layers while maintaining FP32 accumulations, delivers maximum throughput. Researchers anticipate further gains on newer Ampere/Hopper GPUs utilising TF32/BF16, and from kernel-level FP16/BF16 paths and pipeline fusion, paving the way for even more efficient molecular simulations.
Machine Learning Accelerates Molecular Dynamics Simulations
This work demonstrates substantial acceleration of molecular dynamics simulations using machine-learning force fields, specifically the MACE model, without compromising accuracy. Researchers achieved speed improvements by optimising computational bottlenecks and evaluating the use of reduced-precision arithmetic. The team found that employing the cuEquivariance backend reduces inference latency and that casting only linear layers to lower precision formats, such as BF16 or FP16, within an FP32 model yields significant speedups. Importantly, these gains were achieved while maintaining the integrity of energies and thermodynamic observables in both short and long simulations. Investigating the impact of training at lower precision revealed that while it degrades force accuracy, careful implementation of mixed-precision inference, using FP32 for accumulations, can mitigate these effects. The authors acknowledge that further performance gains are likely with newer generation hardware and through additional kernel-level optimisations.
👉 More information
🗞 Speeding Up MACE: Low-Precision Tricks for Equivarient Force Fields
🧠 ArXiv: https://arxiv.org/abs/2510.23621